The rate of methicillin-resistant Staphylococcus aureus (MRSA) among the total of S. aureus isolates decreased to 35.3% in 2017 in China. It is unclear whether the molecular characteristics of S. aureus isolates have changed as the rate decreased.
ObjectiveThis study aimed to investigate the molecular characteristics and virulence genes profile of S. aureus isolates causing bloodstream infection and analyze the correlation between the prevalence rates of the common sequence types and MRSA.
MethodsA total of 112 S. aureus strains from eight hospitals of four cities, including 32 MRSA isolates, were identified and evaluated through multilocus sequence typing, spa typing, and determination of virulence genes.
ResultsTwenty-five STs were identified, of which ST5 (21.4%) was the most prevalent, whereas the prevalence of ST239 correlated with the rate of MRSA among all S. aureus isolates. Forty-six spa types were identified, of which t2460 (14.3%) was the most common. clfa, hla, seb, fnbA and hlb were the prevailing virulence genes. 81.3% MRSA and 45.0% methicillin-sensitive S. aureus (MSSA) isolates harbored six or more tested virulence genes. ST5-t2460, seldom noted in bloodborne S. aureus isolates in China, was the most common clone. The prevalence of harboring six or more virulence genes in ST5-t2460 and ST188-t189 were 93.8% and 8.3%, respectively.
ConclusionST5-t2460 was the most common clone in S. aureus causing bloodstream infection followed by ST188-t189, which had never been noted in China before. Moreover, ST5-t2460 harbored more virulence genes than ST188-t189, and the prevalence of ST239 clone decreased with the proportion of MRSA among all S. aureus isolates.
Staphylococcus aureus (S. aureus) is an important pathogen causing a great number of infectious diseases. Recently, with the development of organ transplantation, bone marrow transplantation, and invasive diagnostic tests, bloodstream infections such as bacteremia and septicemia are on the rise. S. aureus is one of the most common human pathogen causing bloodstream infection, which can lead to both community- and hospital-acquired bacteremia. The mortality of patients with S. aureus bacteremia was reported to be about 30%,1 which was closely associated with methicillin-resistant S. aureus (MRSA) infections.2 Compared to patients with methicillin-sensitive S. aureus (MSSA) bacteremia, the mortality MRSA bacteremia increases by 40%.
It was observed that the molecular characteristics of S. aureus have regional differences. Sequence type 239 (ST239) were found to be the most prevalent in many Asian countries including China,3,4 but ST36 and ST30 were the most frequent in the United Kingdom.5 Researches about molecular characteristics and virulence gene profiles of bloodborne S. aureus isolates began late in China. A study carried out in 2012 which revealed that ST239-t030 and ST188-t189 were the most prevalent among the MRSA isolates, whereas ST7-t091 was the most prevalent among the MSSA isolates followed by ST188-t189 and ST630-t377.6 Subsequent studies showed that ST239-t030 and ST239-t037 were the most common in MRSA isolates, whereas ST7-t091 and ST188-t189 were the most prevalent in MSSA isolates.7–9 MRSA isolates carry more virulence encoding genes than MSSA isolates according to studies indicating that MRSA isolates have different molecular characteristics and virulence gene profiles compared to MSSA isolates.6,8,9
Recently, with the increasing of awareness of infections control, the proportion of MRSA among all S. aureus isolates decreases significantly. China Antimicrobial Surveillance Network (CHINET) showed that the ratio of MRSA was 69.0% in 2005 and 35.3% in 2017.10 Because MRSA isolates differ from MSSA isolates in molecular characteristics and virulence genes profile, it is reasonable to speculate that the molecular characteristics and virulence genes profile of S. aureus isolates change as the proportion of MRSA among all S. aureus isolates decreases. The aims of this study were to investigate the molecular characteristics and virulence genes profile of bloodborne S. aureus, and analyze the correlation between the prevalence of the common STs and the proportion of MRSA among all S. aureus isolates. We also compared the differences of molecular characteristics and virulence genes profile between MRSA and MSSA isolates.
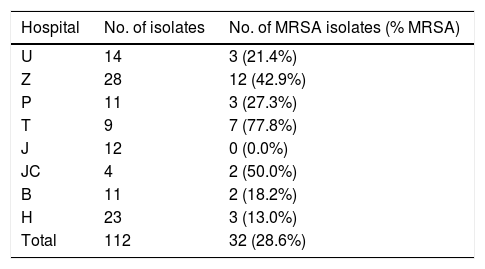
Material and methodsS. aureus isolation, identification and collectionA total of 112 non-duplicate S. aureus isolates were collected from eight hospitals in four cities from July 2017 to February 2018 (see Table 1). Isolates were identified as S. aureus using conventional microbiological methods including Gram staining, catalase and coagulase tests. They were further identified by Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) System (BD, USA). An antimicrobial susceptibility test was first carried out using a VITEK 2 Compact system and VITEK 2 AST-GP67 Test Kit (bioMerieux, Inc., Durham, NC, USA). Methicillin resistance was confirmed using a cefoxitin disk (30μg, Oxoid) and oxacillin disk (1μg, Oxoid,) in Zhongnan Hospital, Wuhan university. All the isolates were stored at −80°C for further experiments. The Ethics Committee of Zhongnan Hospital, Wuhan University approved this study. Because this was a retrospective study and all patients were anonymized, informed consent was waived.
The distribution of 112 S. aureus isolates.
| Hospital | No. of isolates | No. of MRSA isolates (% MRSA) |
|---|---|---|
| U | 14 | 3 (21.4%) |
| Z | 28 | 12 (42.9%) |
| P | 11 | 3 (27.3%) |
| T | 9 | 7 (77.8%) |
| J | 12 | 0 (0.0%) |
| JC | 4 | 2 (50.0%) |
| B | 11 | 2 (18.2%) |
| H | 23 | 3 (13.0%) |
| Total | 112 | 32 (28.6%) |
U, Union Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology; Z, Zhongnan Hospital of Wuhan University; P, Puai Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology; T, Tongji Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology; J, the First People's Hospital of Jingzhou; JC, Jingzhou Central Hospital; B, Beijing Tsinghua Changgung Hospital Affiliated to Tsinghua University; H, Hainan General Hospital.
All S. aureus isolates were grown on blood agar at 37°C overnight, then a single colony was transferred into 5mL Tryptic Soy Broth (TSB) medium to culture for 16h at a rotation speed of 200rpm/min. S. aureus pellets were used to extract chromosomal DNA with TIANamp Bacteria DNA kit (Tiangen, China) supplemented with 1mg/mL lysostaphin (Sigma, China) according to the manufacturer's instructions. All the extracted chromosomal DNAs were stored at −20°C for further tests.
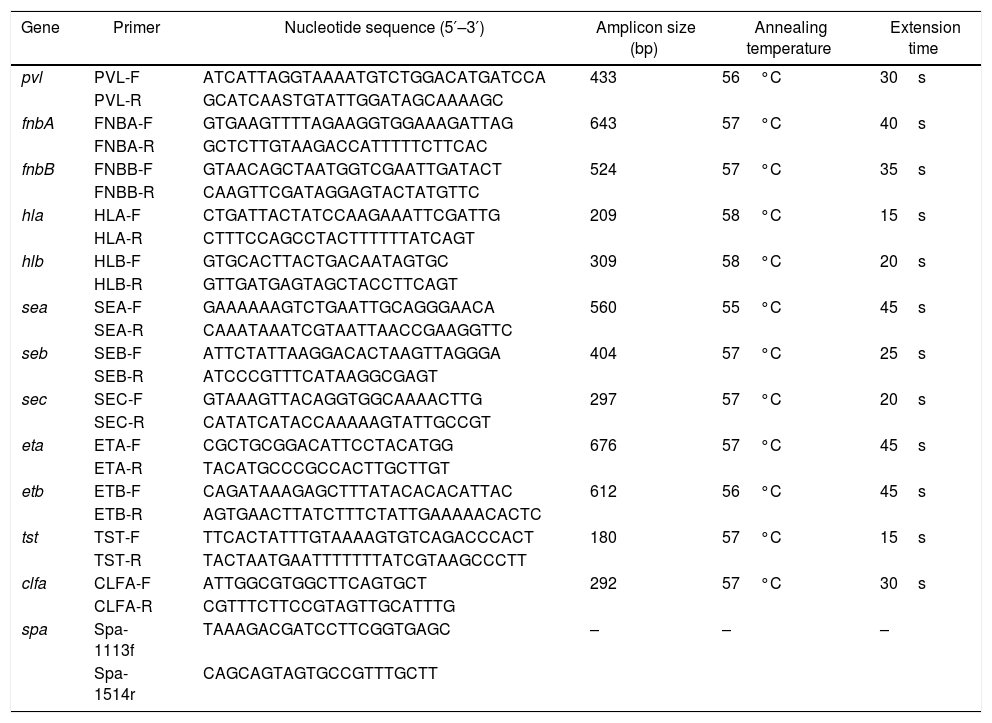
Staphylococcal protein A (spa) typingThe polymorphic X region of spa consists of a variable number of 24bp repeat units allowing isolates to be distinguished from one another. In the present study, the polymorphic X region of spa was amplified from the extracted chromosomal DNAs using primers spa-1113f and spa-1514r11 (see Table 2). The PCR mixture contained 2μL spa-1113f (10μM), 2μL spa-1514r (10μM), 1μL chromosomal DNA template, 25μL 2×Taq Master Mix (Tiangen, China), and 20μL double distilled water. The PCR conditions were as follow: initial denaturation at 94°C for 5min, followed by 30 cycles of denaturation at 94°C for 45s, annealing at 53°C for 60s and extension at 72°C for 90s, and a final extension at 72°C for 10min. The amplicons were sequenced (Tianyihuiyuan, China) and analyzed using the Ridom web server (http://spaserver.ridom.de).
Primers used in this study and the PCR conditions for amplifying virulence genes.
| Gene | Primer | Nucleotide sequence (5′–3′) | Amplicon size (bp) | Annealing temperature | Extension time |
|---|---|---|---|---|---|
| pvl | PVL-F | ATCATTAGGTAAAATGTCTGGACATGATCCA | 433 | 56°C | 30s |
| PVL-R | GCATCAASTGTATTGGATAGCAAAAGC | ||||
| fnbA | FNBA-F | GTGAAGTTTTAGAAGGTGGAAAGATTAG | 643 | 57°C | 40s |
| FNBA-R | GCTCTTGTAAGACCATTTTTCTTCAC | ||||
| fnbB | FNBB-F | GTAACAGCTAATGGTCGAATTGATACT | 524 | 57°C | 35s |
| FNBB-R | CAAGTTCGATAGGAGTACTATGTTC | ||||
| hla | HLA-F | CTGATTACTATCCAAGAAATTCGATTG | 209 | 58°C | 15s |
| HLA-R | CTTTCCAGCCTACTTTTTTATCAGT | ||||
| hlb | HLB-F | GTGCACTTACTGACAATAGTGC | 309 | 58°C | 20s |
| HLB-R | GTTGATGAGTAGCTACCTTCAGT | ||||
| sea | SEA-F | GAAAAAAGTCTGAATTGCAGGGAACA | 560 | 55°C | 45s |
| SEA-R | CAAATAAATCGTAATTAACCGAAGGTTC | ||||
| seb | SEB-F | ATTCTATTAAGGACACTAAGTTAGGGA | 404 | 57°C | 25s |
| SEB-R | ATCCCGTTTCATAAGGCGAGT | ||||
| sec | SEC-F | GTAAAGTTACAGGTGGCAAAACTTG | 297 | 57°C | 20s |
| SEC-R | CATATCATACCAAAAAGTATTGCCGT | ||||
| eta | ETA-F | CGCTGCGGACATTCCTACATGG | 676 | 57°C | 45s |
| ETA-R | TACATGCCCGCCACTTGCTTGT | ||||
| etb | ETB-F | CAGATAAAGAGCTTTATACACACATTAC | 612 | 56°C | 45s |
| ETB-R | AGTGAACTTATCTTTCTATTGAAAAACACTC | ||||
| tst | TST-F | TTCACTATTTGTAAAAGTGTCAGACCCACT | 180 | 57°C | 15s |
| TST-R | TACTAATGAATTTTTTTATCGTAAGCCCTT | ||||
| clfa | CLFA-F | ATTGGCGTGGCTTCAGTGCT | 292 | 57°C | 30s |
| CLFA-R | CGTTTCTTCCGTAGTTGCATTTG | ||||
| spa | Spa-1113f | TAAAGACGATCCTTCGGTGAGC | – | – | – |
| Spa-1514r | CAGCAGTAGTGCCGTTTGCTT |
MLST was carried out as described previously.12 Seven S. aureus housekeeping genes (i.e. arcC, aroE, glpF, gmk, pta, tpi and yqil) were amplified by seven PCR assays followed by sequencing. The sequences of amplicons were compared to the known alleles stored in the MLST database (http://saureus.mlst.net) to identify the sequence type. Clustering of related STs, which were defined as clonal complexes (CCs), was determined using eBURST.
Detection of virulence genesSeveral PCR assays were used to detect the following 12 staphylococcal virulence genes including the pvl genes (lukF/S-PV), the staphylococcal enterotoxin genes (sea, seb, sec), the exfoliative toxin genes (eta, etb), the hemolysin genes (hla, hlb), the adhesion factor genes (fnbA, fnbB, clfa), and the toxic shock syndrome toxin (tst). All primers are listed in Table 2.13–15 The PCR mixture contained 2μL of a couple of primers (10μM), 1μL DNA template, 12.5μL Taq Master Mix (2×), and double distilled water (9.5μL). The PCR conditions were as follow: initial denaturation at 94°C for 5min, followed by 30 cycles of denaturation at 94°C for 30s, 30s with the respective annealing temperature, extension at 72°C for respective extension time, and a final extension at 72°C for 10min. The annealing temperatures and extension times for different primers are shown in Table 2. All PCR products were analyzed by electrophoresis with 2% agarose gels.
Statistical analysisStatistical analyses were performed using SPSS Statistics 24.0 for Windows. The chi-square test was used to compare the distributions of virulence genes between MRSA and MSSA isolates, ST5-t2460 and ST188-t189. The correlations between the prevalence of ST239, ST5 clones and the proportion of MRSA among all S. aureus isolates were analyzed using Spearman rank correlation. p<0.05 was considered to be statistically significant.
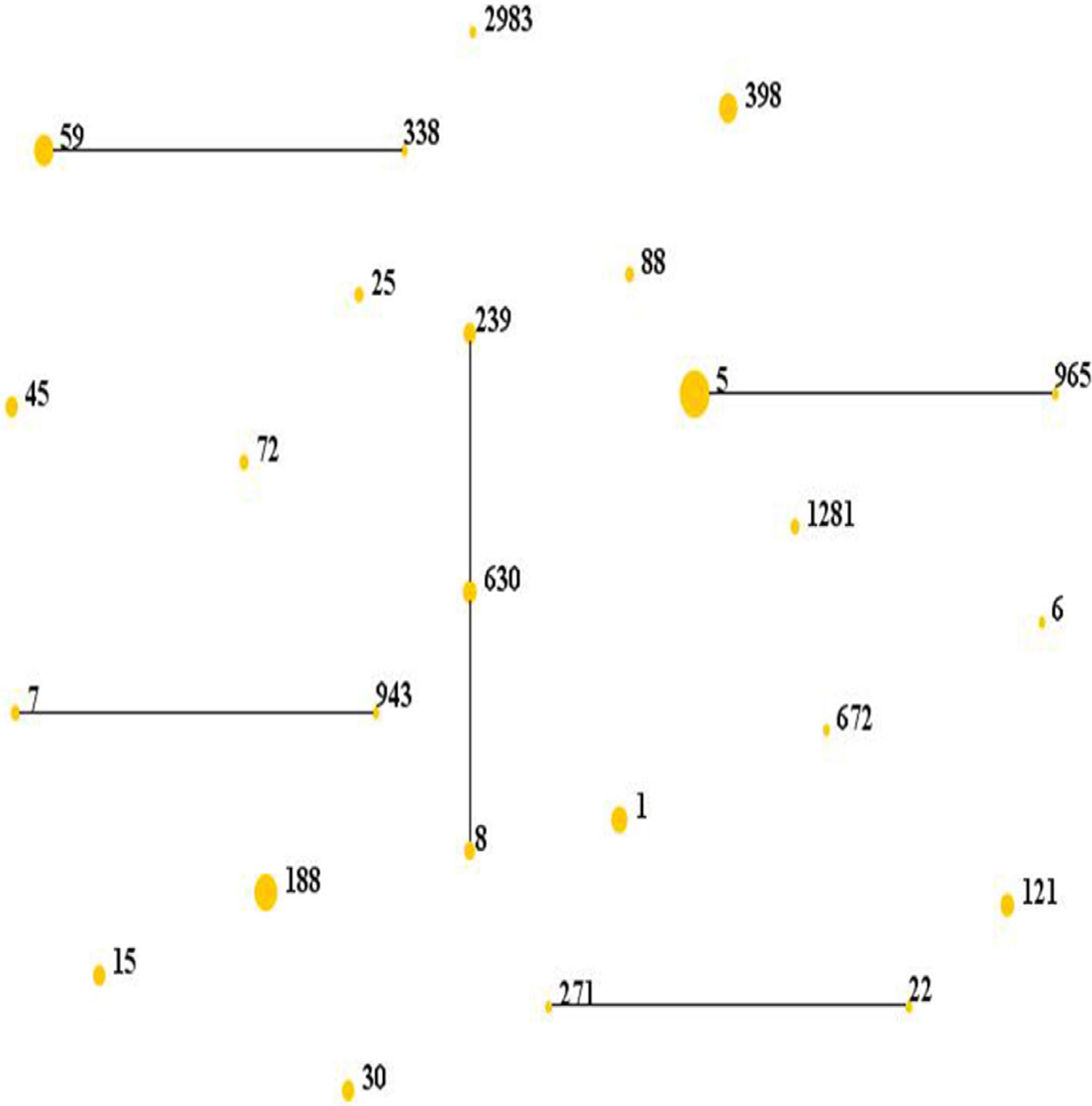
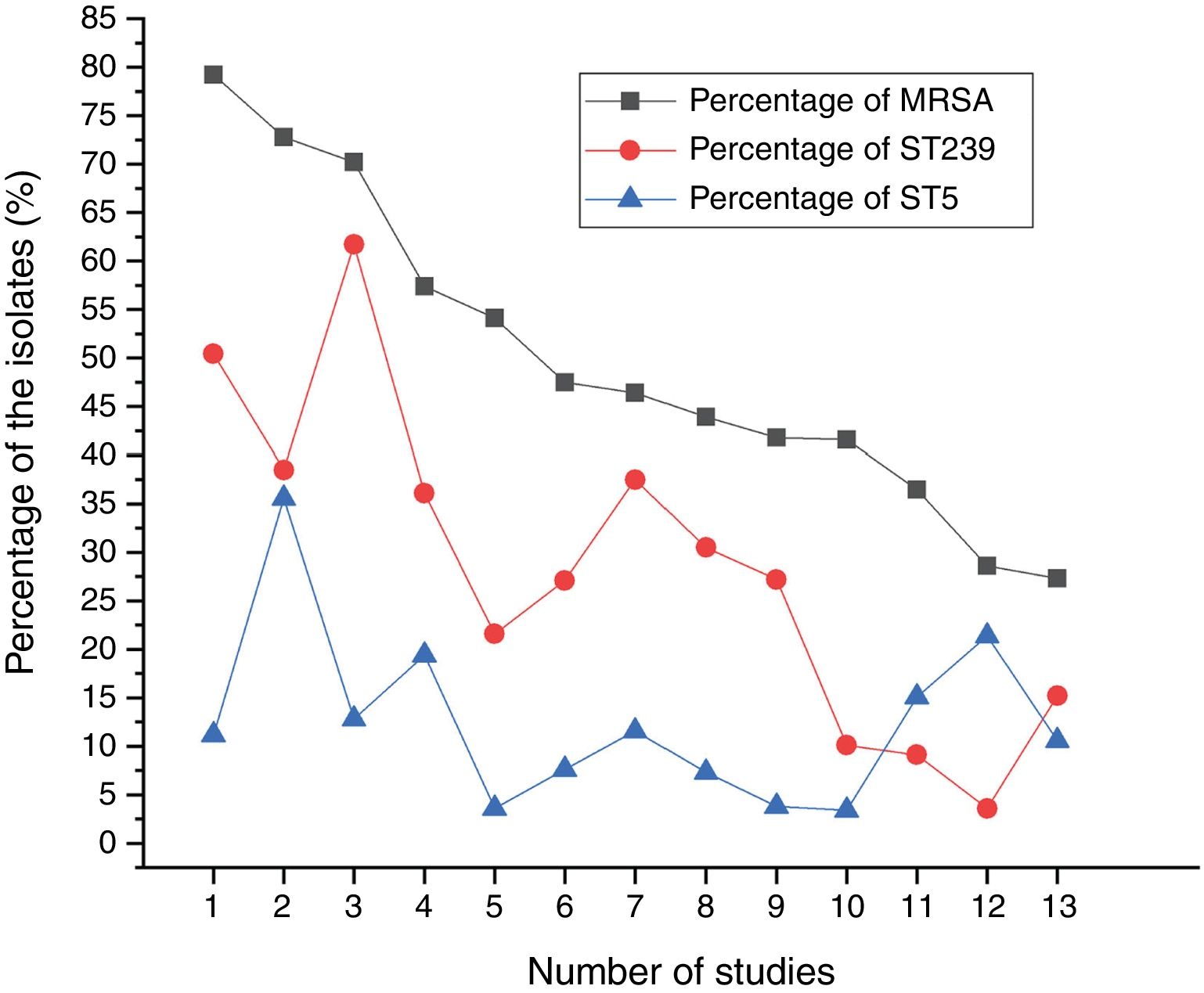
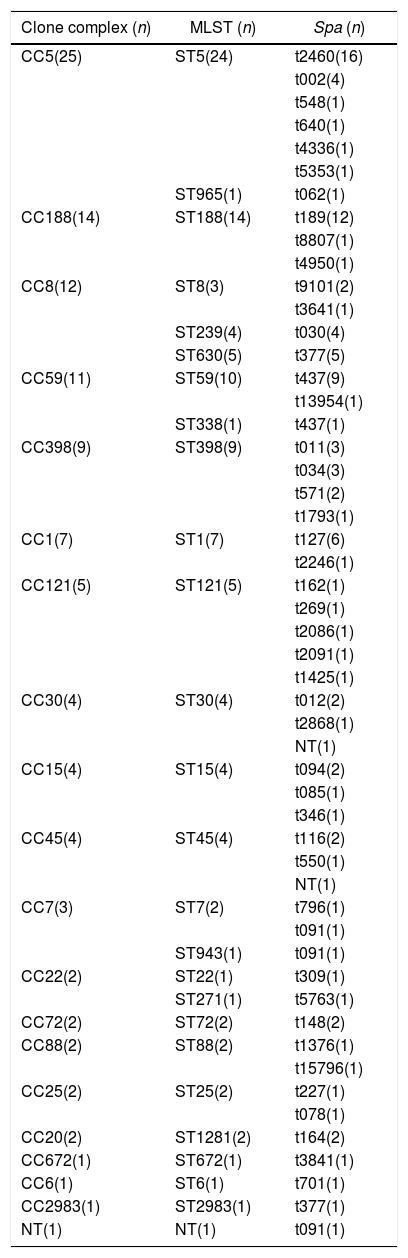
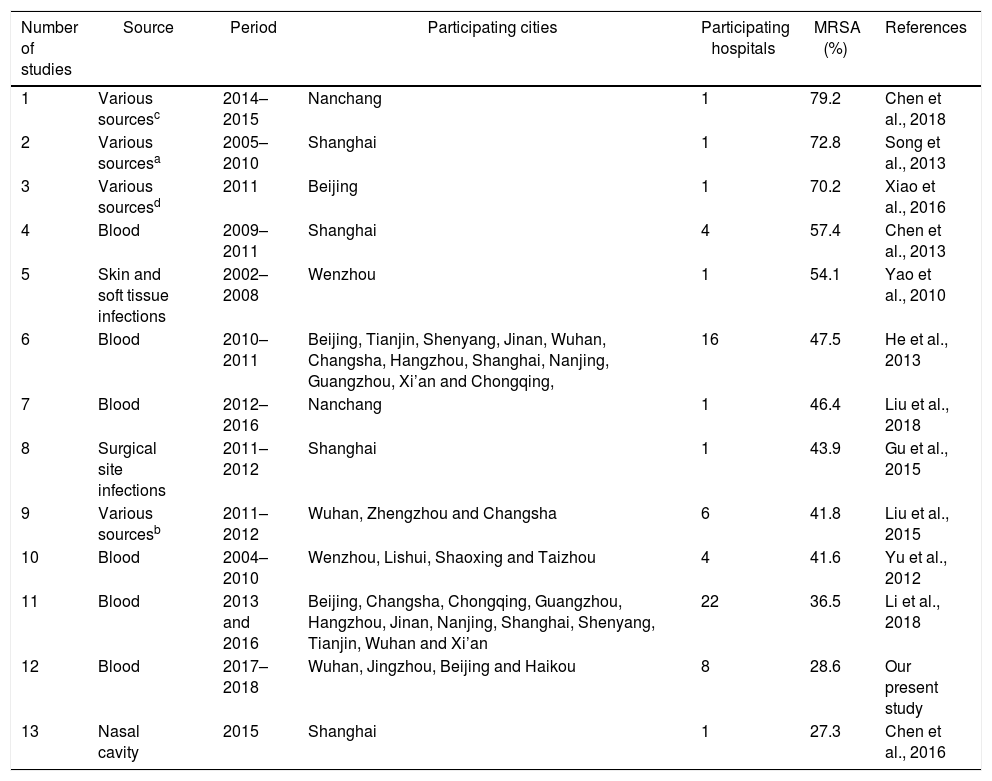
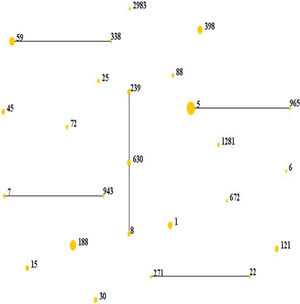
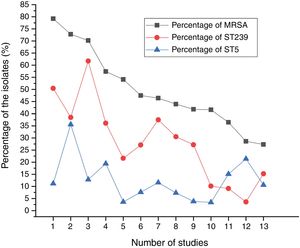
Results and discussionMolecular typing of S. aureus isolates.The distribution of MLST and spa types of bloodborne S. aureus isolates is presented in Table 3. Among the 112 S. aureus isolates, 25 STs were identified. ST5 was the most predominant (21.4%), followed by ST188 (12.5%), ST59 (8.9%), ST398 (8.0%), ST1 (6.3%), ST630 (4.5%), and ST121 (4.5%). Other STs accounted for 33.9% of the 112 S. aureus isolates. In addition, one isolate could not be identified for ST. 95.8% of the ST5 isolates were collected from Wuhan, whereas 57.1% of the ST188 isolates were isolated from Haikou, which indicates that the distribution of STs of S. aureus have regional differences. Sixteen clone complexes (CCs) and three singletons were identified by eBURST (Fig. 1), CC5 (22.3%) was the most prevalent clonal complex, followed by CC8 (10.7%) and CC59 (9.8%). Thirty-two (28.6%) S. aureus isolates were MRSA in this study. The most major epidemic ST in MRSA isolates was ST5 (50.0%), followed by ST59 (15.6%), ST239 (12.5%), and ST45 (9.4%). In contrast, the most prevalent ST in MSSA isolates was ST188 (17.5%) followed by ST398 (10.0%) and ST5 (10.0%). In last ten years in China, five studies about molecular characteristics of S. aureus isolates causing bloodstream infection were carried out.6,8,9,16,17 ST239 was found to be the most prevalent clone except in the studies by Yu and Li where ST239 was the most common clone among the MRSA isolates, suggesting that the prevalence of ST239 clone may be closely associated with MRSA infections. Twelve previous studies about molecular characteristic profiles of S. aureus isolates in China6,8,9,16–24 and our present study analyzed the correlation between the proportion of MRSA among all S. aureus isolates and the prevalence of ST239 and ST5 clone. All S. aureus isolates both MRSA and MSSA in the above studies were collected from patients rather than volunteers. The information of these S. aureus isolates is listed in Table 4. The rates of MRSA to total S. aureus isolates and the prevalence of ST239 and ST5 clone are shown in Fig. 2. Spearman rank correlation analysis showed that the proportion of MRSA among all S. aureus isolates correlated closely with the prevalence of ST239 clone (r=0.8, p<0.001) rather than ST5 clone (r=0.2, p=0.5), and the prevalence of ST239 clone decreased as the proportion of MRSA among all S. aureus isolates decreased, indicating that molecular characteristics of S. aureus isolates change as the proportion of MRSA among all S. aureus isolates fall. These studies also indicated that the implementation of infection control contributed to preventing the spread of ST239 S. aureus isolates.
Molecular characteristics of S. aureus isolates from patients with bloodstream infection.
| Clone complex (n) | MLST (n) | Spa (n) |
|---|---|---|
| CC5(25) | ST5(24) | t2460(16) |
| t002(4) | ||
| t548(1) | ||
| t640(1) | ||
| t4336(1) | ||
| t5353(1) | ||
| ST965(1) | t062(1) | |
| CC188(14) | ST188(14) | t189(12) |
| t8807(1) | ||
| t4950(1) | ||
| CC8(12) | ST8(3) | t9101(2) |
| t3641(1) | ||
| ST239(4) | t030(4) | |
| ST630(5) | t377(5) | |
| CC59(11) | ST59(10) | t437(9) |
| t13954(1) | ||
| ST338(1) | t437(1) | |
| CC398(9) | ST398(9) | t011(3) |
| t034(3) | ||
| t571(2) | ||
| t1793(1) | ||
| CC1(7) | ST1(7) | t127(6) |
| t2246(1) | ||
| CC121(5) | ST121(5) | t162(1) |
| t269(1) | ||
| t2086(1) | ||
| t2091(1) | ||
| t1425(1) | ||
| CC30(4) | ST30(4) | t012(2) |
| t2868(1) | ||
| NT(1) | ||
| CC15(4) | ST15(4) | t094(2) |
| t085(1) | ||
| t346(1) | ||
| CC45(4) | ST45(4) | t116(2) |
| t550(1) | ||
| NT(1) | ||
| CC7(3) | ST7(2) | t796(1) |
| t091(1) | ||
| ST943(1) | t091(1) | |
| CC22(2) | ST22(1) | t309(1) |
| ST271(1) | t5763(1) | |
| CC72(2) | ST72(2) | t148(2) |
| CC88(2) | ST88(2) | t1376(1) |
| t15796(1) | ||
| CC25(2) | ST25(2) | t227(1) |
| t078(1) | ||
| CC20(2) | ST1281(2) | t164(2) |
| CC672(1) | ST672(1) | t3841(1) |
| CC6(1) | ST6(1) | t701(1) |
| CC2983(1) | ST2983(1) | t377(1) |
| NT(1) | NT(1) | t091(1) |
NT, non-typeable; n, number of isolates in each type. The spa typing was based on variations of the repeat units, amplification and sequencing of the X region were performed as described previously by Strommenger et al.11 MLST of S. aureus isolates was performed using detection of the 7 housekeeping genes of S. aureus as described previously.12
Distribution of STs in the clonal complexes. The diagram generated by eBURST with the default group definition based on the MLST data of this study, representing the relationships of 111 S. aureus isolates. Each number implies an MLST ST, STs that are linked by a line belong to the same cluster and the dot area indicates the prevalence of the ST in the MLST data of this study.
Desc ription of the S. aureus isolates included in 12 previous studies and our present study.
| Number of studies | Source | Period | Participating cities | Participating hospitals | MRSA (%) | References |
|---|---|---|---|---|---|---|
| 1 | Various sourcesc | 2014–2015 | Nanchang | 1 | 79.2 | Chen et al., 2018 |
| 2 | Various sourcesa | 2005–2010 | Shanghai | 1 | 72.8 | Song et al., 2013 |
| 3 | Various sourcesd | 2011 | Beijing | 1 | 70.2 | Xiao et al., 2016 |
| 4 | Blood | 2009–2011 | Shanghai | 4 | 57.4 | Chen et al., 2013 |
| 5 | Skin and soft tissue infections | 2002–2008 | Wenzhou | 1 | 54.1 | Yao et al., 2010 |
| 6 | Blood | 2010–2011 | Beijing, Tianjin, Shenyang, Jinan, Wuhan, Changsha, Hangzhou, Shanghai, Nanjing, Guangzhou, Xi’an and Chongqing, | 16 | 47.5 | He et al., 2013 |
| 7 | Blood | 2012–2016 | Nanchang | 1 | 46.4 | Liu et al., 2018 |
| 8 | Surgical site infections | 2011–2012 | Shanghai | 1 | 43.9 | Gu et al., 2015 |
| 9 | Various sourcesb | 2011–2012 | Wuhan, Zhengzhou and Changsha | 6 | 41.8 | Liu et al., 2015 |
| 10 | Blood | 2004–2010 | Wenzhou, Lishui, Shaoxing and Taizhou | 4 | 41.6 | Yu et al., 2012 |
| 11 | Blood | 2013 and 2016 | Beijing, Changsha, Chongqing, Guangzhou, Hangzhou, Jinan, Nanjing, Shanghai, Shenyang, Tianjin, Wuhan and Xi’an | 22 | 36.5 | Li et al., 2018 |
| 12 | Blood | 2017–2018 | Wuhan, Jingzhou, Beijing and Haikou | 8 | 28.6 | Our present study |
| 13 | Nasal cavity | 2015 | Shanghai | 1 | 27.3 | Chen et al., 2016 |
Among the 112 S. aureus isolates, 48 spa types were found. The most prevalent spa type was t2460 (14.3%) followed by t189 (10.7%), t437 (8.9%), t377 (5.4%), t127 (5.4%), t002 (3.6%), and t030 (3.6%). In addition, two isolates could not be identified for spa. All the t2460 S. aureus isolates were collected from Wuhan except one isolate from Beijing, whereas the t189 S. aureus isolates were collected in Haikou (58.3%), Wuhan (25.0%), Jingzhou (8.3%) and Beijing (8.3%). In 2007, t2460 clone was first found in South Korea,25 whereas in China it was not be found in large numbers until May 2015.26 These data may supported the view of the temporary outbreak of t2460 in Wuhan. To date, t091 and t030 were the most common spa types in bloodstream infection in China,6–9 whereas t437 and t127 were found to be the most common spa types in other infections.27–29
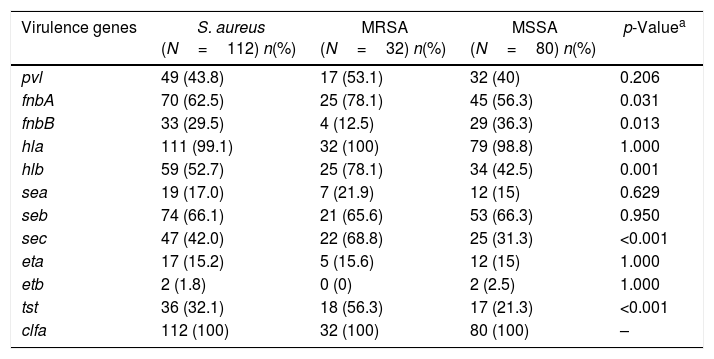
Virulence gene profilesThe pathogenicity of S. aureus is closely related to presence of various virulence genes.30 The frequencies of virulence encoding genes identified in the 112 isolates are listed in Table 5. Our data showed that all S. aureus isolates carried at least two virulence genes and almost all the isolates contained clfa (100%, 112/112) and hla (99.1%, 111/112) genes. These findings are comparable to those reported in many hospitals of south of China,31,32 which indicated that clfa and hla were widely present and may play a pivotal role in the pathogenicity of S. aureus. Enterotoxins, including sea, seb and sec were found to cause food poisoning primarily. In our study, the most common enterotoxin gene was seb (66.1%), which is significantly different from those reported in foodborne S. aureus, where the most common enterotoxin gene is sea.33,34 Our study showed that 55.4% (62/112) isolates harbored six or more virulence genes, of which two isolates contained nine virulence genes, 15 isolates harbored eight virulence genes, 15 isolates carried eight virulence genes, 18 isolates had seven virulence genes, 27 isolates harbored six virulence genes. Furthermore, six or more virulence genes were present in 81.3% of MRSA isolates and 45% of MSSA isolates and MRSA isolates harbored more virulence genes than the MSSA isolates (χ2=12.154, p<0.01). The above results suggested that S. aureus isolates with different genetic background have different ability to acquire mobile genetic elements such as plasmids, phages and pathogenicity islands.
The frequencies of virulence encoding genes among S. aureus, MRSA and MSSA isolates in bloodstream infection.
| Virulence genes | S. aureus (N=112) n(%) | MRSA (N=32) n(%) | MSSA (N=80) n(%) | p-Valuea |
|---|---|---|---|---|
| pvl | 49 (43.8) | 17 (53.1) | 32 (40) | 0.206 |
| fnbA | 70 (62.5) | 25 (78.1) | 45 (56.3) | 0.031 |
| fnbB | 33 (29.5) | 4 (12.5) | 29 (36.3) | 0.013 |
| hla | 111 (99.1) | 32 (100) | 79 (98.8) | 1.000 |
| hlb | 59 (52.7) | 25 (78.1) | 34 (42.5) | 0.001 |
| sea | 19 (17.0) | 7 (21.9) | 12 (15) | 0.629 |
| seb | 74 (66.1) | 21 (65.6) | 53 (66.3) | 0.950 |
| sec | 47 (42.0) | 22 (68.8) | 25 (31.3) | <0.001 |
| eta | 17 (15.2) | 5 (15.6) | 12 (15) | 1.000 |
| etb | 2 (1.8) | 0 (0) | 2 (2.5) | 1.000 |
| tst | 36 (32.1) | 18 (56.3) | 17 (21.3) | <0.001 |
| clfa | 112 (100) | 32 (100) | 80 (100) | – |
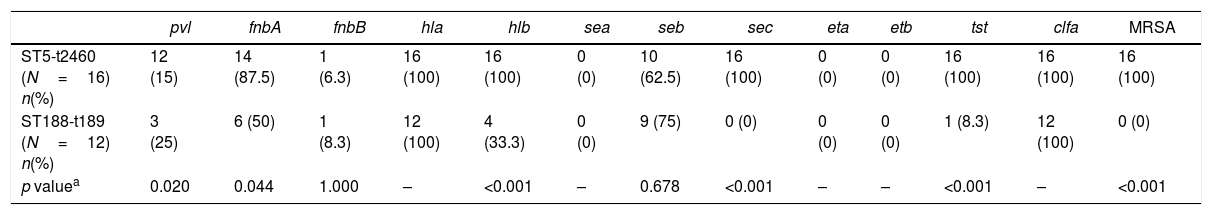
MLST typing and spa typing were performed to analyze the molecular characteristics of S. aureus isolates. There was a strong association observed between ST and spa types: the ST5 type was primarily associated with t2460 (66.7%, 16/24), the ST188 type with t189 (85.7%, 12/14), the ST59 type with t437 (90%, 9/10), the ST1 type with t127 (85.7%, 6/7) and the ST630 type with t377 (100%, 5/5). ST5-t2460 (16/112, 14.3%) were found to be the most common in this study, followed by ST188-t189 (12/112, 10.7%). Molecular characteristics of ST5-t2460 and ST188-t189 are summarized in Table 6. 93.8% of ST5-t2460 were collected from four hospitals in Wuhan, whereas the distribution of ST188-t189 was more dispersed including in Haikou (58.3%), Wuhan (25%), Beijing (8.3%) and Jingzhou (8.3%). In 2007, ST5-t2460 became the main type of MRSA bloodstream infections in Korea which had never been seen before.25 In China, extremely few cases of ST5-t2460 were reported in previous studies.16,35 Therefore, the prevalence of ST5-t2460 in our research may be consequence of a temporary outbreak of ST5-t2460 in Wuhan. In our present study, all the ST5-t2460 were MRSA, whereas all the ST188-t189 were MSSA. The prevalence of strains with six or more virulence genes was significantly higher in ST5-t2460 strains (93.8%) compared with ST188-t189 strains (8.3%) (p<0.001). In addition, of the tested 12 virulence genes, pvl, fnbA, hlb, sec, and tst were found to be more frequent in ST5-t2460 than in ST188-t189. These results indicated that ST5-t2460 acquire mecA and virulence genes easily, which supported the above conclusion that S. aureus with different genetic background have different ability to acquire mobile genetic elements. In addition to genetic background, ST5-t2460 often lead to persistent and recurrent MRSA bloodstream infections,36 which also contribute to promoting S. aureus isolates to acquire virulence genes easily.
The frequencies of virulence encoding genes among S. aureus, ST5-t2460 S. aureus and ST188-t189 S. aureus isolates in bloodstream infection.
| pvl | fnbA | fnbB | hla | hlb | sea | seb | sec | eta | etb | tst | clfa | MRSA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST5-t2460 (N=16) n(%) | 12 (15) | 14 (87.5) | 1 (6.3) | 16 (100) | 16 (100) | 0 (0) | 10 (62.5) | 16 (100) | 0 (0) | 0 (0) | 16 (100) | 16 (100) | 16 (100) |
| ST188-t189 (N=12) n(%) | 3 (25) | 6 (50) | 1 (8.3) | 12 (100) | 4 (33.3) | 0 (0) | 9 (75) | 0 (0) | 0 (0) | 0 (0) | 1 (8.3) | 12 (100) | 0 (0) |
| p valuea | 0.020 | 0.044 | 1.000 | – | <0.001 | – | 0.678 | <0.001 | – | – | <0.001 | – | <0.001 |
In conclusion, ST5-t2460 was the most common clone in S. aureus causing bloodstream infection, followed by ST188-t189, which had never been reported in China before. All the ST5-t2460 were MRSA, whereas all the ST188-t189 were MSSA. Moreover, ST5-t2460 harbored more virulence genes than ST188-t189, which indicates that ST5-t2460 acquire mecA and virulence genes more easily than ST188-t189. In addition, the prevalence of ST239 clone decreased with the decrease of the proportion of MRSA among all S. aureus isolates.
FundingThis study was supported by the National Natural Science Foundation of China (Grant No. 81371779) and the Natural Science Foundation of Hubei Province (Grant No. 2016CFB672).
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to thank all the hospitals for providing clinical isolates and data.