The aim of this study was to perform SCCmec typing in Staphylococcus aureus isolates and to characterize the clonal profile of these isolates. Forty-six mecA gene-positive strains isolated between 2002 and 2006 were submitted to antimicrobial resistance testing by the E-test, SCCmec typing by multiplex PCR, and clonal profile analysis by pulsed-field gel electrophoresis. Forty-one (89.1%) isolates were typed as SCCmec III and five (10.9%) as SCCmec IV. Four circulating clones were detected, one of them comprising isolates related to the Brazilian epidemic clone. This clone was detected throughout the study period. The SCCmec III isolates were associated with a high rate of multidrug resistance and clonal dissemination of methicillin-resistant S. aureus in the wards of the University Hospital of the Botucatu School of Medicine, Universidade Estadual Paulista.
Staphylococcus aureus is a major human pathogen, which has been related to numerous pathological processes, such as food poisoning, pneumonia, bacteremia, impetigo, folliculitis, and osteomyelitis.1 At present, this microorganism is the main causative agent of nosocomial infections. In Brazil, approximately 20% of bacteremias are caused by S. aureus.2 Similar rates have been reported in several European countries, indicating that it is important to study the role of this microorganism in healthcare-associated infections.3,4
Oxacillin is used as a resistance marker for methicillin-resistant S. aureus (MRSA), since strains resistant to oxacillin are often multiresistant. Approximately 30–50% of hospital S. aureus isolates are oxacillin-resistant, a fact that leaves few treatment options.5–7 The mecA gene is the main mechanism responsible for methicillin resistance. This gene is carried by the cassette chromosome mec (SCCmec), a movable genetic element that consists of incision and excision genes (ccr complex), in addition to genes that encode resistance to antimicrobials and heavy metals.8 Eleven different SCCmec types have been described so far,9 which differ in terms of the number of genes that they carry and gene architecture. In addition, different SCCmec types are associated with hospital or community isolates, a fact making SCCmec typing an important epidemiological tool.10 Some of these SCCmec types carry genes encoding multidrug resistance, including resistance to beta-lactams, macrolides, lincosamines, streptogramins, aminoglycosides, and tetracycline. Hence, if a bacterial cell acquires such SCCmec, it concomitantly acquires a multidrug resistance phenotype.11
In Brazil, oxacillin resistance rates are high, with an incidence of 30–50% depending on the region studied.12–14 However, little is known about the dissemination of these clonal isolates, although studies on the clonal profile of MRSA infections have been conducted in other countries. Five large pandemic clones of MRSA isolates have been identified by epidemiological techniques. Recent findings permitted us to confirm important changes in the epidemiology of MRSA infections in Brazilian hospitals. At the beginning of the past decade, the Brazilian epidemic clone (BEC)/type III/ST239 was the main lineage in Brazilian hospitals.15 Over recent years, type IV isolates have emerged in hospital settings and have become a common type of MRSA found in inpatients worldwide. Studies conducted in Rio de Janeiro, Brazil, identified USA400/type IV/ST1 followed by USA800/ST5 as the main lineages.16–19 Antimicrobial resistance and virulence factors are unique to each clone.
Since oxacillin resistance has become a major problem in hospitals which leaves few therapeutic options and since the presence of pandemic clones indicates clonal dissemination of MRSA isolates, the aim of the present study was to perform molecular characterization of MRSA strains isolated from the University Hospital of the Botucatu School of Medicine (HCFMB), UNESP.
Forty-six mecA gene-positive S. aureus strains (MRSA) isolated from blood cultures of patients hospitalized in different wards of HCFMB-UNESP between 2002 and 2006 and stored in the culture collection of the Department of Microbiology and Immunology, UNESP, were studied. The strains were isolated using the standard microbiological techniques described by Koneman et al.20 In this study, the isolates of the 30 HCFMB-UNESP wards were divided into complexes according to specialties: Internal Medicine Wards, Surgery Wards, Pediatric Wards, Gynecology and Obstetrics, Emergency Room, Intensive Care Units, and other wards.
The in vitro susceptibility of the isolates to the following drugs was tested: oxacillin, netilmicin, erythromycin, sulfamethoxazole-trimethoprim, and vancomycin. For this purpose, the minimal inhibitory concentration (MIC) of these drugs was determined by the E-test. The isolates were classified as sensitive or resistant according to the cut-off levels (μg/mL) recommended by the CLSI.21
SCCmec typing was performed by multiplex PCR as described by Milheiriço et al.22 The S. aureus isolates were typed by PFGE according to the modified protocol of McDougal et al.23 Among the 46 mecA gene-positive S. aureus isolates, 23 were selected based on a similar profile of MIC determined with the E-test strip. The isolates were divided into groups according to the MIC obtained for all antimicrobial agents tested and one isolate representative of each group was selected. All S. aureus strains carrying SCCmec type IV were submitted to typing by PFGE. SmaI (Fast Digest SmaI, Fermentas Life Science, Canada) was used for genomic DNA restriction. Electrophoresis was performed on 1% agarose gel prepared with 0.5M TBE (Pulsed Field Certified Agarose, BioRad Laboratories, USA) using the CHEF-DR III System (BioRad Laboratories) under the following conditions: pulse time interval of 5–40s for 21h on a linear ramp; 6V/cm; 120° angle; 14° C; 0.5M TBE as running buffer. Lambda Ladder PFG Marker (New England BioLabs, UK) was used as a molecular weight marker. The gels were stained with GelRed (10,000× in water, Biotium, USA) for 1h and photographed under UV transillumination. For similarity analysis, the Dice correlation coefficient was estimated and a dendrogram was created by the UPGMA method (unweighted pair group method using arithmetic averages) using the BioNumerics 6.1 software (Applied Maths, Belgium). Band position tolerance and optimization were set at 1.5 and 1%, respectively. A similarity coefficient of 80% was chosen for cluster definition.
Among the 46 mecA gene-positive S. aureus strains isolated from blood cultures between 2002 and 2006, 41 (89.1%) were typed as SCCmec III and five (10.9%) as SCCmec IV. Most (n=14) of the 41 SCCmec III isolates were identified in the Internal Medicine complex, followed by nine isolates in other wards, eight isolates in the Surgery complex, seven isolates in the Emergency complex, two isolates in the Intensive Care Unit complex, and one isolate in the Nursery complex. SCCmec type IV was detected in two isolates from the Emergency complex and in one isolate each from the Surgery, Internal Medicine and Intensive Care Unit complexes.
Resistance to the drugs tested varied according to SCCmec type. A higher percentage of isolates resistant to erythromycin (n=40), netilmicin (n=38) and trimethoprim–sulfamethoxazole (n=39) was observed among SCCmec III MRSA. An association was observed between multidrug resistance and presence of the SCCmec III cassette, with 38 (92.7%) isolates being resistant to three of the four drugs tested and only one isolate being resistant to two, one and none of the drugs, respectively. Four of the five SCCmec IV isolates were susceptible to the drugs tested and one strain was resistant to erythromycin. SCCmec III MRSA predominated in all years studied; however, SCCmec IV isolates were not detected in 2005.
Susceptibility to vancomycin varied according to SCCmec cassette type. The MIC50 and MIC90 of vancomycin for SCCmec IV isolates were 2 and 3μg/mL, respectively. These values were higher than those obtained for SCCmec III isolates (1.5 and 2μg/mL, respectively). No decrease in vancomycin susceptibility was observed in SCCmec III and IV isolates over the study period. Among SCCmec III isolates, the MIC50 of vancomycin showed a slight decrease from 2002 (2μg/mL) to 2003 (1μg/mL), followed by a slight increase in 2004 (1.5μg/mL), and remained stable in 2005 and 2006 (2μg/mL). For SCCmec IV isolates, MIC values decreased from 2002 to 2003 and remained stable in subsequent years.
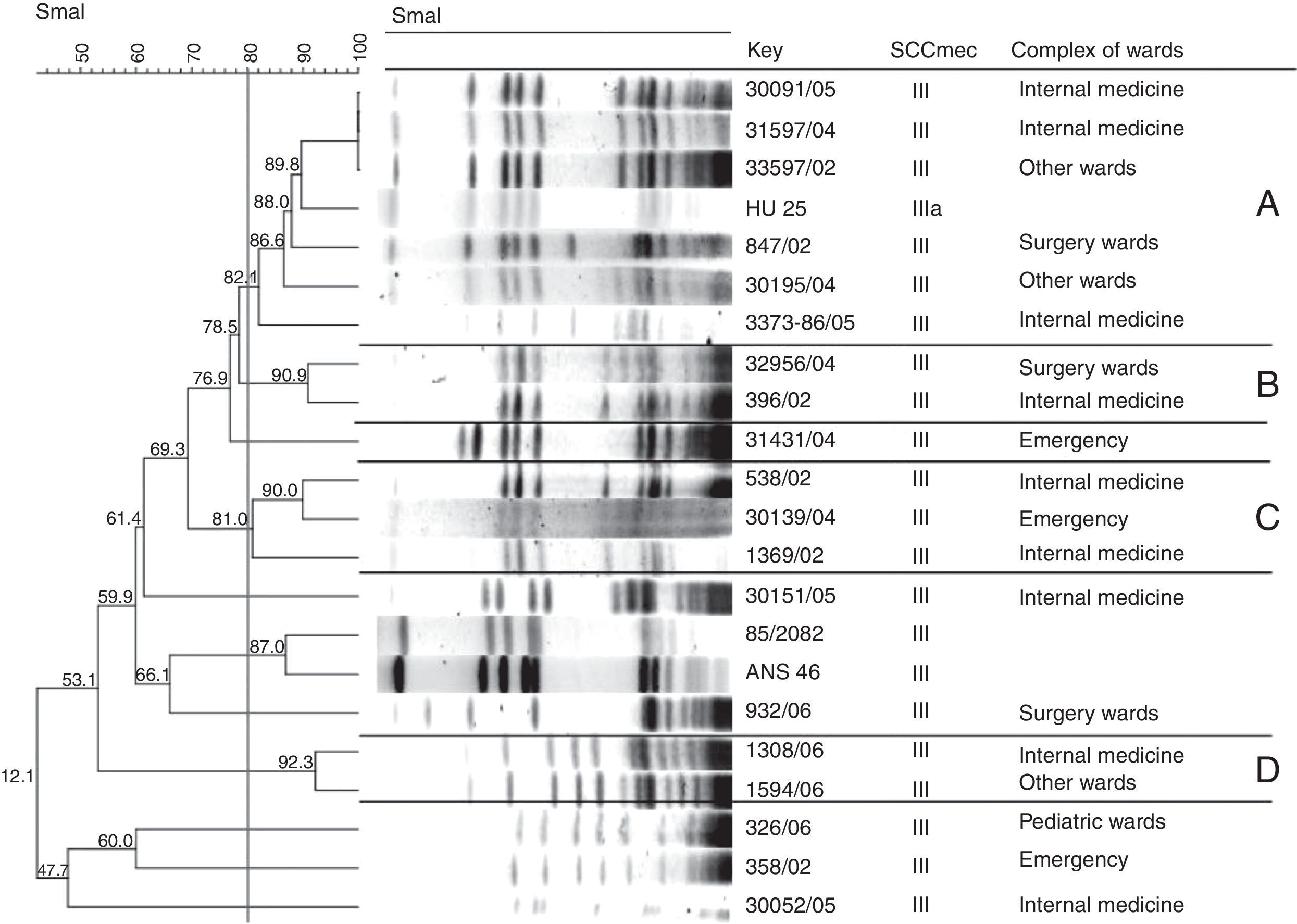
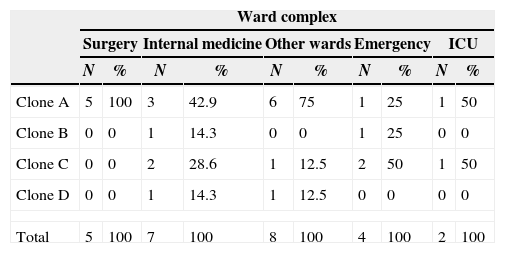
Analysis of SCCmec III isolates by PFGE revealed the presence of four clones. These clones were divided based on a similarity coefficient ≥80% and identified with capital letters. There was a predominance of clone A, comprising five isolates. When all isolates were analyzed, 16 S. aureus strains isolated from the Internal Medicine, Surgery, Emergency and other wards complexes were included in this clone (Table 1 and Fig. 1). This clone predominated in the Surgery and other wards complexes.
Distribution of clones of MRSA isolates carrying SCCmec type III according to ward complex.
| Ward complex | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Surgery | Internal medicine | Other wards | Emergency | ICU | ||||||
| N | % | N | % | N | % | N | % | N | % | |
| Clone A | 5 | 100 | 3 | 42.9 | 6 | 75 | 1 | 25 | 1 | 50 |
| Clone B | 0 | 0 | 1 | 14.3 | 0 | 0 | 1 | 25 | 0 | 0 |
| Clone C | 0 | 0 | 2 | 28.6 | 1 | 12.5 | 2 | 50 | 1 | 50 |
| Clone D | 0 | 0 | 1 | 14.3 | 1 | 12.5 | 0 | 0 | 0 | 0 |
| Total | 5 | 100 | 7 | 100 | 8 | 100 | 4 | 100 | 2 | 100 |
Clone: identified in the dendrogram of SCCmec type III Staphylococcus aureus generated by Dice/UPGMA analysis (Bionumerics, Applied Maths) of PFGE profiles (SmaI) (similarity ≥80%).
N: number of MRSA isolates belonging to the respective clone.
Surgery complex: surgery, cardiac surgery, thoracic surgery, gastric surgery, orthopedic surgery, and urologic surgery wards.
Internal medicine: dermatology, gastrointestinal clinic, internal medicine, intensive care unit, cardiology, cardiology clinic, hematology and pulmonology wards.
Other wards: chemotherapy, oncology, infectious and parasitic diseases, neurology, and dialysis wards.
Emergency: emergency room and emergency room-intensive care unit.
ICU: intensive care unit including intensive care unit, central intensive care unit and coronary intensive care unit.
Dendrogram of SCCmec type III Staphylococcus aureus isolates generated by Dice/UPGMA analysis (Bionumerics, Applied Maths) of PFGE profiles (SmaI) (similarity≥80%) and comparison with international MRSA clones harboring SCCmec type III. The international clones were kindly provided by Dr. Antonio Carlos Campos Pignatari, Laboratório Especial de Microbiologia Clínica, Disciplina de Infectologia, Universidade Federal de São Paulo-Escola Paulista de Medicina, and Dr. Agnes Marie Sá Figueiredo, Instituto de Microbiologia Prof. Paulo de Góes, Universidade Federal do Rio de Janeiro, Brazil.
The reference strain HU 25 CEB, which belongs to the Brazilian clone of pandemic circulation (BEC), was also included in clone A, suggesting a relationship between these isolates and the Brazilian epidemic clone. Strains isolated between 2002 and 2006 were included in this clone (Fig. 1). Clone B comprised two strains isolated from the Internal Medicine and Emergency complexes between 2002 and 2004 (Table 1 and Fig. 1). Clone C comprised three isolates (Fig. 1). However, analysis of MIC by the E-test resulted in the inclusion of six strains isolated in 2002, 2004, 2005 and 2006 from the Intensive Care Unit, Emergency and other wards complexes in clone C, with a higher prevalence in the Internal Medicine and Emergency complexes (Table 1 and Fig. 1). Clone D comprised only two strains isolated in 2006 from the Internal Medicine and other wards complexes (Table 1 and Fig. 1). Twenty isolates exhibited a polyclonal profile.
The SCCmec IV isolates showed low clonality and only two strains isolated in 2002 and 2004 from the Internal Medicine and Intensive Care Unit complexes could be clustered.
The present study showed a predominance of the SCCmec III cassette in S. aureus. This fact emphasizes the importance of appropriate antibiotic therapy since nosocomial Staphylococcus spp. isolates are known to be associated with multidrug resistance. There was a strong association between SCCmec III isolates and multidrug resistance, which can be explained by the nosocomial origin of these isolates. The drug of choice for the treatment of infections with multiresistant MRSA and S. aureus is vancomycin; however, vancomycin-resistant isolates or isolates with intermediate susceptibility have been identified. There are also studies describing decreased susceptibility of hospital isolates to vancomycin.24,25 Decreased vancomycin susceptibility was not observed in the present study, with the MIC50 and MIC90 values of SCCmec III MRSA isolates being stable over the study period. However, an association between the presence of SCCmec III and isolates with intermediate vancomycin resistance has been reported.26
Analysis of the prevalence of SCCmec cassette types in MRSA isolates over the study period showed an increase in the number of SCCmec III from 2002 to 2005, followed by a decrease in 2006. The prevalence of SCCmec IV was low and stable between 2002 and 2004 and this type was detected again only in 2006.
In view of the increase in the number of oxacillin-resistant Staphylococcus spp. and the emergence of heteroresistant and vancomycin-resistant isolates, studies investigating the dissemination of these isolates in hospitals are needed. PFGE has been used successfully for this purpose. In the present study, 23 S. aureus isolates were typed by this method. Although a high degree of clonality has been reported for Brazilian nosocomial MRSA,27 only four clones were detected in the present study, with the circulation of isolates in various wards and prevalence during the studied years. Clone A comprised the largest number of SCCmec III isolates, which were isolated between 2002 and 2006 from the following ward complexes: Surgery, Internal Medicine, other wards, Emergency, and Intensive Care Unit. The HU 25 strain, which belongs to BEC, showed a similarity of more than 80% to clone A. Hence, all isolates belonging to clone A are related to BEC. The Brazilian clone predominates in hospitals in all regions of Brazil, in South America and in some European countries such as Portugal, accounting for 70% of MRSA isolated from hospitals.15 A high prevalence of BEC-like isolates was detected in hospitals in the northeastern and southeastern regions of Brazil.28,29 In the present study, isolates belonging to clone A were identified in all years studied, a finding that can be explained by the fact that clone A has been the predominant clone in various Brazilian hospitals since the 1990s.29,30 Although the clone related to BEC has been the most predominant, its prevalence was not homogeneous, since other clones were found in this study, which can mean a replacement in the future circulation of the BEC clone by other clones.
The isolates characterized as SCCmec IV showed a polyclonal profile. However, PFGE permitted the identification of a clone that disseminated in 2002 and 2004. Isolates of this clone were recovered from the Cardiology and Intensive Care Unit wards. This clone did not show similarity to strains USA800 or HAR24, which belong to the pediatric and EMRSA-15 clone, respectively, and are characterized by the presence of the SCCmec IV cassette. Although SCCmec IV isolates did not predominate in this study, replacement of BEC with EMRSA-15 has been reported in Portugal.31 A high percentage of SCCmec IV MRSA isolates (46%) was described in a Swiss university hospital, with the predominance of four major clones. This finding can be explained by the replacement of hospital clones with pediatric clones in healthcare-associated infections or by a massive prevalence of isolates of community origin.32 In Brazil, three strains genetically related to clone USA800 were isolated in the northeastern region, a finding suggesting the introduction of this community-acquired clone into hospital settings.28 Studies conducted in Rio de Janeiro suggest that, in Brazil, USA400/type IV isolates of community origin have gradually replaced BEC/type III in the hospital environment.16–19 However, only one SCCmec IV S. aureus strain isolated from the Emergency ward showed a similarity of 60.9% with this clone, indicating that they are different clones. The high prevalence of BEC isolates in the present study might be explained by the collection of S. aureus strains included in the study (2002–2006). Further studies need to be conducted at this hospital to determine whether a change is occurring in the clonal profile of isolates.
The present results confirm the importance of the epidemiological characterization of MRSA isolates considering the high rates of multidrug resistance among nosocomial S. aureus isolates and clonal dissemination of MRSA isolates in the wards of HCFMB-UNESP.
FundingThis study was supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).
Conflicts of interestThe authors declare no conflicts of interest.