Several in-house PCR-based assays have been described for the detection of bacterial meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae from clinical samples. PCR-based methods targeting different bacterial genes are frequently used by different laboratories worldwide, but no standard method has ever been established. The aim of our study was to compare different in-house and a commercial PCR-based tests for the detection of bacterial pathogens causing meningitis and invasive disease in humans.
MethodsA total of 110 isolates and 134 clinical samples (99 cerebrospinal fluid and 35 blood samples) collected from suspected cases of invasive disease were analyzed. Specific sets of primers frequently used for PCR-diagnosis of the three pathogens were used and compared with the results achieved using the multiplex approach described here. Several different gene targets were used for each microorganism, namely ctrA, crgA and nspA for N. meningitidis, ply for S. pneumoniae, P6 and bexA for H. influenzae.
ResultsAll used methods were fast, specific and sensitive, while some of the targets used for the in-house PCR assay detected lower concentrations of genomic DNA than the commercial method. An additional PCR reaction is described for the differentiation of capsulated and non-capsulated H. influenzae strains, the while commercial method only detects capsulated strains.
ConclusionsThe in-house PCR methods here compared showed to be rapid, sensitive, highly specific, and cheaper than commercial methods. The in-house PCR methods could be easily adopted by public laboratories of developing countries for diagnostic purposes. The best results were achieved using primers targeting the genes nspA, ply, and P6 which were able to detect the lowest DNA concentrations for each specific target.
Infections caused by Neisseria meningitidis (Nm), Streptococcus pneumoniae (Sp) and Haemophilus influenzae (Hi) are responsible for high morbidity and mortality rates among children and adults in many countries each year.1,2 Despite effective antimicrobial and supportive therapy, mortality rates among children remain high, with significant long-term sequelae in survivors. Although adequate treatment requires rapid detection and identification of the bacteria, traditional laboratory diagnostic methods such as culture for the identification of bacterial meningitis pathogens may take 36h or more. Because of the similarity of clinical symptoms among invasive infections caused by Nm, Sp and Hi, physicians must frequently request laboratories to test the clinical sample for these three microorganisms to determine the causative bacteria and decide on the management of the case. Rapid diagnosis of the etiological agent is also important for health surveillance, in cases of invasive disease, to avoid transmission to close contacts.1–3
To address this problem, nonculture methods like PCR have become available during the last two decades, providing early and accurate diagnosis of bacterial meningitis.1,4–10 Because of its rapid, sensitive and specific results, PCR is an important tool to improve the diagnostic of such microorganisms. An additional advantage of PCR over conventional laboratory methods is the possibility to detect genomic DNA from clinical samples without the need of previous culture. Recently, the use of a quantitative PCR method called Real Time-PCR, have improved the diagnosis of infectious agents based on molecular methods.11–17 However, the higher cost of reagents and equipment could be a possible drawback to the implementation of this method in public laboratories of developing countries. The use of a basic PCR method, can be still more appropriate for diagnostic purposes in low-budget laboratories. A number of different target genes have been used in several PCR-based assays to specifically detect the genomic DNA of these agents. For N. meningitidis nspA, ctrA and crgA, for H. influenzae bexA and P6, and for S. pneumoniae ply and lytA genes have been used as targets.4–10,18,19 In order to determine the best combination of primers, we have developed a simple multiplex PCR for the simultaneous detection of the three main agents of meningitis worldwide, Nm, Sp, and Hi. The multiplex PCR method here described allows the detection of the three microorganisms in a single-tube reaction, saving time and reagents. Target species specific genes used for Nm, Hi, and Sp detection were nspA, P6, and ply respectively. Primers amplifying higher length fragments were chosen for identification because the visualization of bands below 200bp through electrophoresis may not be clear depending of the agarose gel type and concentration used. Furthermore, we have compared the specificity and sensitivity of our method to other in-house and commercial PCR-based methods previously described for simultaneous or single detection of Nm, Hi, and Sp from clinical samples.4,5,7–10,18,19
Materials and methodsBacterial isolates, clinical samples, and culture conditionsWe have analyzed a total of 110 strains isolated from clinical samples (N. meningitidis n=31, H. influenzae n=29, S. pneumoniae n=50) which are part of the Bacterial Culture Collection of the INCQS/FIOCRUZ institute. The strains were isolated during the last five years, from clinical samples of cerebrospinal fluid (CSF) and blood. These samples have been collected from culture-confirmed cases of patients with suspected meningitis in Brazilian public hospitals. The isolates were recovered from clinical samples after inoculation in chocolate agar (blood agar base with 5% sterile defribinated rabbit blood at 56°C) and incubated at 37°C in a 5% CO2 environment. The isolated strains were identified as Nm, Hi, or Sp by colony morphology, latex agglutination of specific antisera (Slidex Meningitis – Biomerieux), Gram staining and optochin susceptibility, the latter only for Sp determination. Each species was confirmed by specific PCR tests8,18,19 freeze-dried and incorporated into the culture collection.
We have also analyzed 134 clinical samples (99 CSF and 35 blood) collected from suspected cases of invasive disease. Thirty four (25.3%) of these samples were considered positive for one of the three pathogens targeted in this study by conventional laboratory tests (culture, microscopy, and latex agglutination) and 100 samples were considered negative after conventional laboratory tests, but collected from patients with suspected symptoms or epidemiological link to invasive disease caused by one of the three agents here described. The study has been approved by the local research ethics committee.
Twenty nine reference strains were used as positive controls as follows: N. meningitidis serogroup A (ATCC 13077), N. meningitidis serogroup B (ATCC 13090), N. meningitidis serogroup C (ATCC 13102), N. meningitidis serogroup W135 (ATCC 35559), H. influenzae (ATCC 33391), H. influenzae aegyptius (ATCC 11116), H. influenzae NT (ATCC 49247), H. influenzae serotype a (ATCC 9006), H. influenzae serotype b (ATCC 33533) H. influenzae serotype c (ATCC 9007), H. influenzae serotype d (ATCC 9008), H. influenzae serotype e (ATCC 8142), H. influenzae serotype f (ATCC 9833), S. pneumoniae (ATCC 33400), S. pneumoniae serotype 14 (ATCC 6314), S. pneumoniae serotype 3 (ATCC 6303), S. pneumoniae serotype 33 (ATCC 8333), S. pneumoniae serotype 41 (ATCC 10341), S. pneumoniae serotype 51 (ATCC 10351), S. pneumoniae serotype 19F (ATCC 49619), S. pneumoniae serotype 61 (ATCC 10361), S. pneumoniae serotype 19A (ATCC 700673), S. pneumoniae serotype 14 (ATCC 700672), S. pneumoniae serotype 9V (ATCC 700671), S. pneumoniae serotype 6B (ATCC 700670), S. pneumoniae serotype 4 (ATCC BAA-334), S. pneumoniae serotype 6A (ATCC BAA-659), S. pneumoniae serotype 5 (ATCC BAA-341). Other 9 reference strains were also used as negative controls as they can also cause bacterial meningitis in humans: Neisseria lactamica (ATCC 23970), Neisseria subflava (ATCC 11076), Moraxella catarrhalis (ATCC 25238), Streptococcus agalactiae (ATCC 13813), Streptococcus pyogenes (ATCC 19615), Klebsiella pneumoniae (ATCC 13883), Listeria monocytogenes (ATCC 15313), Acinetobacter sp. (ATCC 14293), Escherichia coli (ATCC 11775).
Extraction and purification of bacterial DNAGenomic DNA extraction and purification from isolated strains and negative clinical samples was carried out using the DNeasy Blood and Tissue extraction kit (Qiagen) according to manufacturer instructions. Briefly, 200μl of the clinical sample were processed using the above mentioned extraction kit, which allows total purification of bacterial genomic DNA free of contaminants and enzyme inhibitors. Extraction and purification of DNA was achieved using different washing buffers and separation on Anion-Exchange Resin columns by binding of nucleic acids and further elution for final use. Purified DNA were kept at −20°C for repeated experiments.
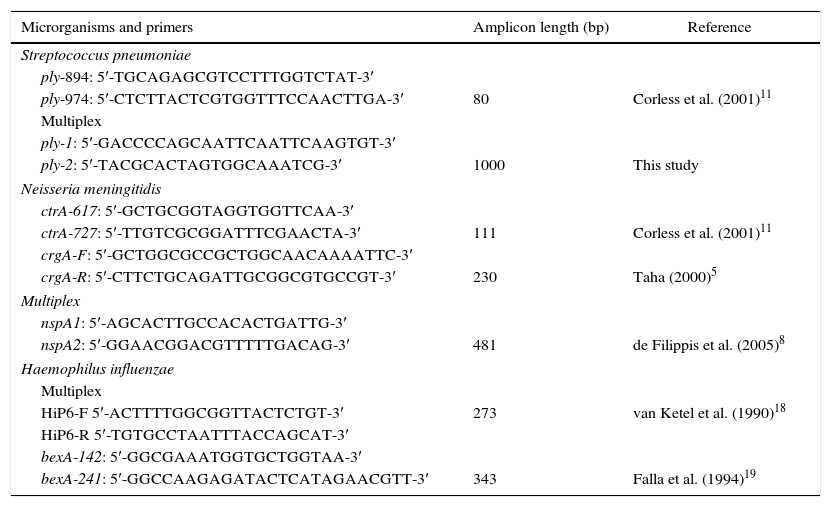
Primer design and PCR conditionsSpecific sets of primers frequently used for PCR-diagnosis of the three above-mentioned agents were used and compared with the results achieved using the multiplex approach here described (Table 1). For the detection of Nm by PCR, target-genes ctrA, crgA and nspA were used, for Sp detection two different regions of the target-gene ply were used. Primers ply-894 and ply-974 were described by Corless et al.,11 targeting a specific region of the pneumococcus pneumolysin gene (ply) amplifying a 80bp amplicon. A new pair of primers was designed to amplify a larger segment of the ply gene. We used the S. pneumoniae pneumolysin gene GenBank access #M17717 sequence as template for primers design. The region amplified was located between positions 555 and 1554 of the ply gene M17717. For Hi detection, target genes P6 and bexA were used (Table 1). PCR was carried out using the PCR-MasterMix (Promega) DNA amplification kit according to manufacturer specifications with an equimolar mixture of the four primer pairs at 50pmol/μl each for the multiplex approach. PCR was run on an Eppendorf Mastercycler EP thermocycler with 35 cycles of 95°C for 30s, 50°C for 1min, 72°C for 1min and 30s and a final extension of 72°C for 10min. A 10μl sample of the completed reaction mixture was run in a 1% agarose gel stained with ethidium bromide. Amplified products were visualized and photographed under UV light. We have also used a commercial kit for the detection of the three agents targeted in this study, named SPEED-OLIGO BACTERIAL MENINGITIS from Vircell which is a PCR-based method coupled to a dipstick device that enables specific detection of H. influenzae, N. meningitides, and S. pneumoniae in cerebrospinal fluid samples. The reactions were carried out according to the producer instructions.
Primers sequences used in this study.
| Microrganisms and primers | Amplicon length (bp) | Reference |
|---|---|---|
| Streptococcus pneumoniae | ||
| ply-894: 5′-TGCAGAGCGTCCTTTGGTCTAT-3′ | ||
| ply-974: 5′-CTCTTACTCGTGGTTTCCAACTTGA-3′ | 80 | Corless et al. (2001)11 |
| Multiplex | ||
| ply-1: 5′-GACCCCAGCAATTCAATTCAAGTGT-3′ | ||
| ply-2: 5′-TACGCACTAGTGGCAAATCG-3′ | 1000 | This study |
| Neisseria meningitidis | ||
| ctrA-617: 5′-GCTGCGGTAGGTGGTTCAA-3′ | ||
| ctrA-727: 5′-TTGTCGCGGATTTCGAACTA-3′ | 111 | Corless et al. (2001)11 |
| crgA-F: 5′-GCTGGCGCCGCTGGCAACAAAATTC-3′ | ||
| crgA-R: 5′-CTTCTGCAGATTGCGGCGTGCCGT-3′ | 230 | Taha (2000)5 |
| Multiplex | ||
| nspA1: 5′-AGCACTTGCCACACTGATTG-3′ | ||
| nspA2: 5′-GGAACGGACGTTTTTGACAG-3′ | 481 | de Filippis et al. (2005)8 |
| Haemophilus influenzae | ||
| Multiplex | ||
| HiP6-F 5′-ACTTTTGGCGGTTACTCTGT-3′ | 273 | van Ketel et al. (1990)18 |
| HiP6-R 5′-TGTGCCTAATTTACCAGCAT-3′ | ||
| bexA-142: 5′-GGCGAAATGGTGCTGGTAA-3′ | ||
| bexA-241: 5′-GGCCAAGAGATACTCATAGAACGTT-3′ | 343 | Falla et al. (1994)19 |
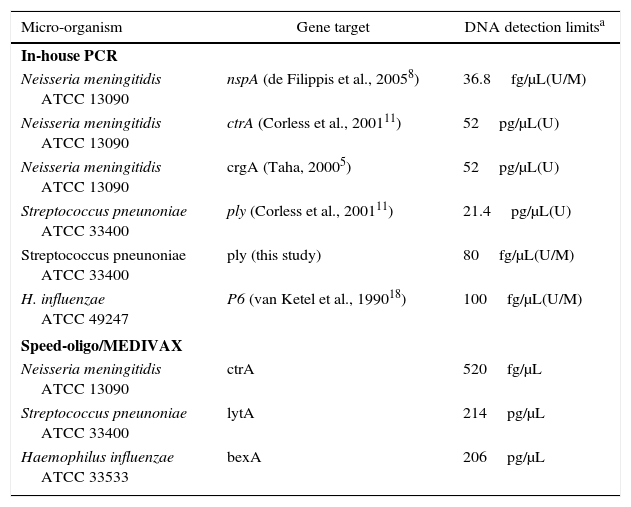
Purified DNA of three reference strains: Nm ATCC 13090, Hi ATCC 33533, 49247, and Sp ATCC 33400 were diluted to an undetectable level for sensitivity determination. Tenfold serial dilutions ranging from 10μg to 10fg DNA per μl were used in single and multiplex PCR giving a range of different sensitivity according to the specific set of primers used (Table 2). Considering a genome size of roughly 2Mb for Nm, Sp, and Hi and since 1kb of ds DNA has a molecular mass of 660kDa, 10fg of DNA represent five genomes.
Sensitivity of in-house-PCR and commercial PCR-based detection kit.
| Micro-organism | Gene target | DNA detection limitsa |
|---|---|---|
| In-house PCR | ||
| Neisseria meningitidis ATCC 13090 | nspA (de Filippis et al., 20058) | 36.8fg/μL(U/M) |
| Neisseria meningitidis ATCC 13090 | ctrA (Corless et al., 200111) | 52pg/μL(U) |
| Neisseria meningitidis ATCC 13090 | crgA (Taha, 20005) | 52pg/μL(U) |
| Streptococcus pneunoniae ATCC 33400 | ply (Corless et al., 200111) | 21.4pg/μL(U) |
| Streptococcus pneunoniae ATCC 33400 | ply (this study) | 80fg/μL(U/M) |
| H. influenzae ATCC 49247 | P6 (van Ketel et al., 199018) | 100fg/μL(U/M) |
| Speed-oligo/MEDIVAX | ||
| Neisseria meningitidis ATCC 13090 | ctrA | 520fg/μL |
| Streptococcus pneunoniae ATCC 33400 | lytA | 214pg/μL |
| Haemophilus influenzae ATCC 33533 | bexA | 206pg/μL |
The differentiation of capsulated and non-capsulated H. influenzae strains was carried out by the inclusion of bexA and P6 primer sets in a second multiplex assay. The presence of two bands at 343bp and 273bp indicated a capsulated Hi strain. With this result an additional PCR is required for serotype determination. The presence of a single band at 273bp indicates a non-capsulated Hi strain and no further serotype determination was necessary.
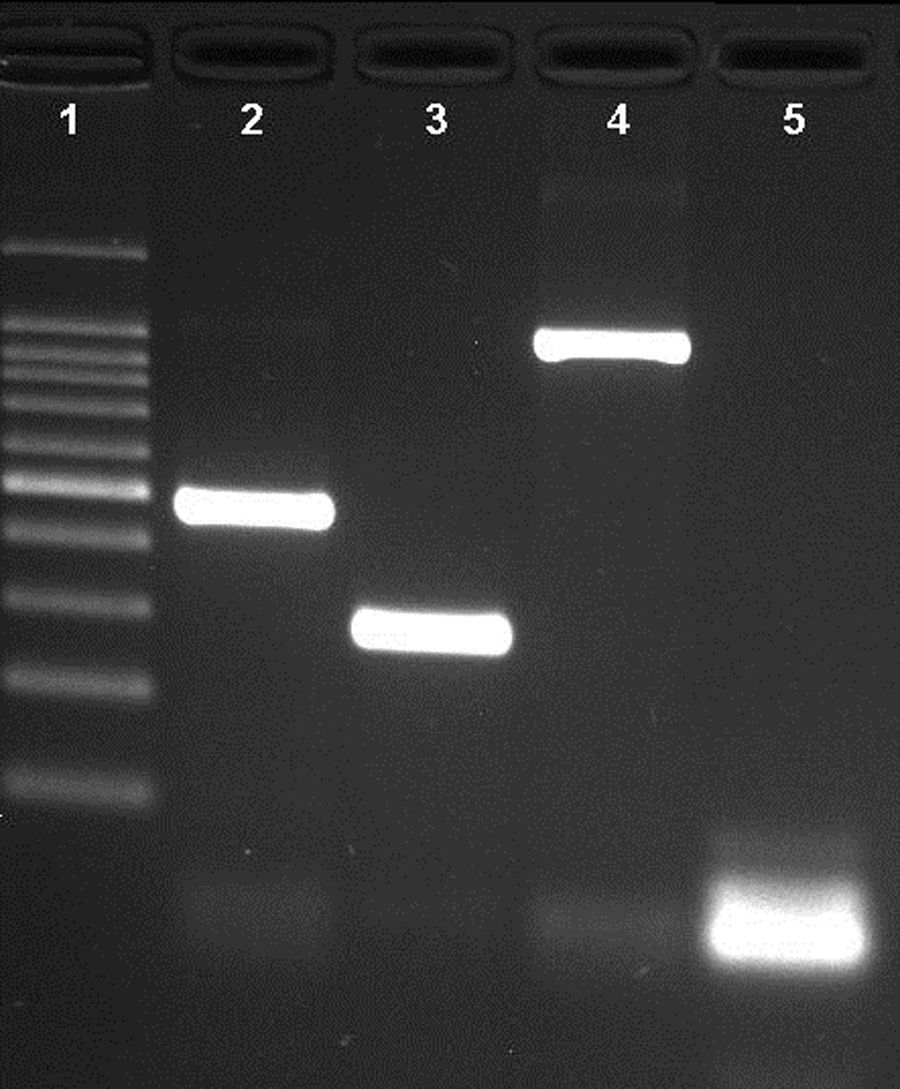
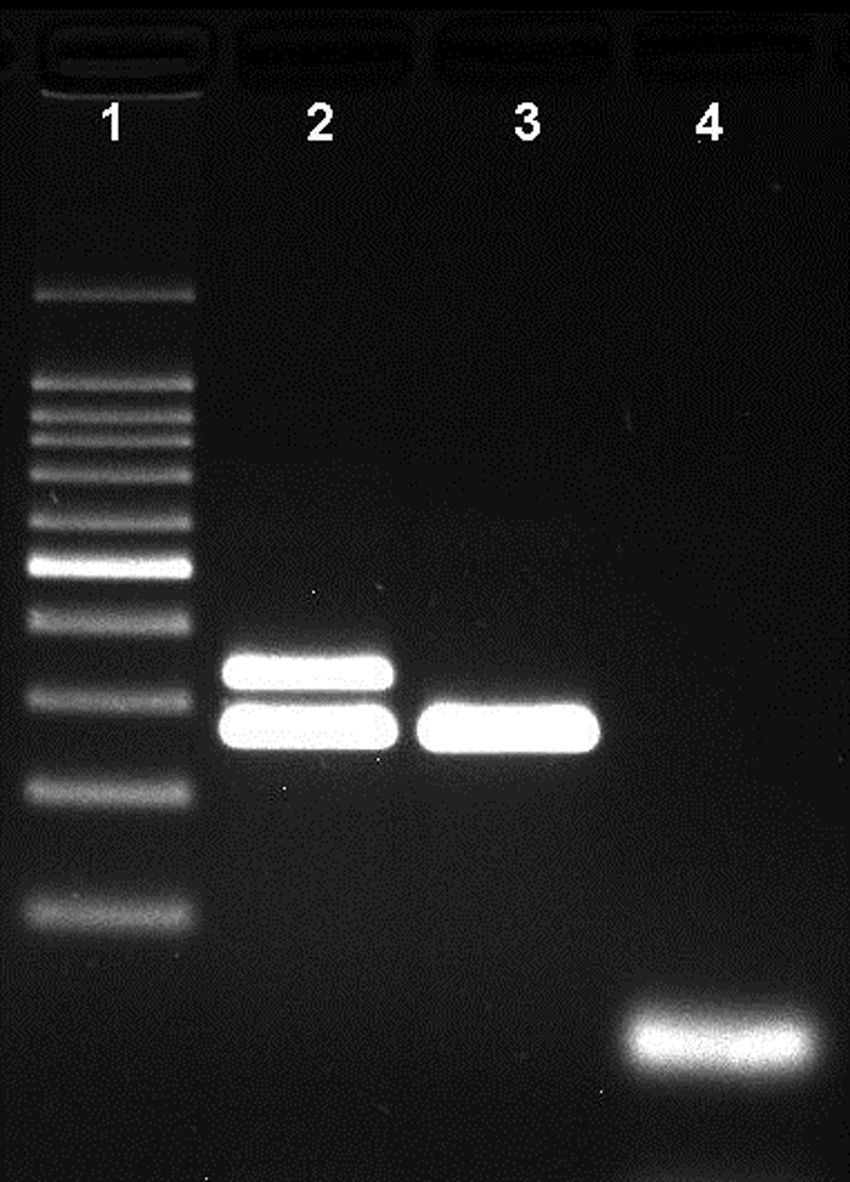
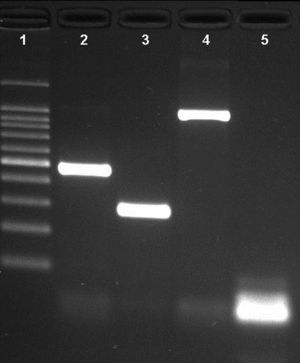
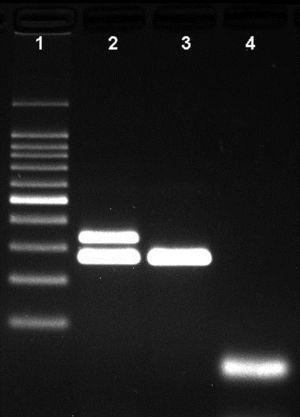
ResultsNovel multiplex PCRThe multiplex PCR approach here described is capable to simultaneously detect genomic DNA of the three agents above described. The three primer pairs used for specifically indicate the etiological agent are nspA (Nm), P6 (Hi), and ply (Sp) giving bands with different sizes: 481bp, 273bp, and 1000bp, respectively (Fig. 1). When a positive result is achieved for Hi, an additional PCR is carried out in order to determine if the strain is capsulated. For this second PCR round, specific primers for targeting the bexA gene are used. Amplification of a 343bp DNA fragment, confirms the presence of a capsulated strain which should be further analyzed to determine the serotype. No amplification after the second PCR round, indicates a non-typeable Hi (HiNT) (Fig. 2).
Agarose gel of a multiplex PCR showing the detection of the three pathogens from three different samples. Lane 1: 100bp DNA ladder; lane 2: Neisseria meningitidis (nspA gene with 481bp); lane 3: Haemophilus influenzae (P6 gene with 273bp); lane 4: Streptococcus pneumoniae (ply gene with 1000bp); lane 5: negative control.
Agarose gel showing the differentiation of capsulated and non-capsulated Haemophilus influenzae strains. Lane 1: 100bp DNA ladder; lane 2: capsulated H. influenzae (bexA and P6 genes with 343bp and 273bp respectively); lane 3: non-capsulated H. influenzae (HiNT), (bexA gene with 343bp); lane 4 negative control.
The specificity of the assay was assessed by testing a panel of non-Nm, Sp, or Hi strains, which included DNAs from nine pathogenic bacteria which may cause invasive disease with symptoms similar to those caused by the three target organisms. No amplification was observed after PCR with any of the microorganisms tested.
SensitivityUniplex and multiplex PCR using the primer sets and the commercial kit here described, showed positive amplification for at least one of the three targets with all the 110 clinical isolates of Nm, Sp, and Hi. For CSF and blood samples, the sensitivity of PCR for detecting bacterial meningitis or septicemia was able to detect bacterial genomic DNA of all the 34 samples previously scored as positive and for additional 12 samples which were scored as negative after conventional laboratory tests giving a total of 46 (34.3%) PCR positive samples (Table 3).
Comparison of detection limits of single, multiplex, and commercial PCR methodsWe have compared different primer sets for the detection of the three microorganisms on single and multiplexed reactions. PCR was carried out as a single or multiplexed reaction performed with the sets of primers described in Table 1. For the commercial kit, the reactions were performed only as a single test (uniplex) for each pathogen as recommended by the producer. As it is shown in Table 2, conventional PCR test using primers targeting genes nspA,8ply (this study) and P618 were able to detect the lowest DNA dilutions of each specific target (36.8fg/μL, 80fg/μL and 100fg/μL, respectively) both in the uniplex or multiplex approach. The commercial test SPEED OLIGO targeting genes ctrA, lytA, and bexA showed to be less sensitive than the conventional PCR test (Table 2).
DiscussionConventional PCR is still widely used as a straightforward laboratory test to detect specific DNA of different pathogens such as virus, bacteria, and protozoa from human or animal clinical samples. Several conventional PCR-based tests have been described for single or simultaneous detection of the most frequent bacterial agents causing invasive disease and meningitis in humans, namely N. meningitidis, S. pneumoniae, and H. influenzae.4–10 Gene targets vary according to the microbial agent showing similar sensitivity and specificity. There are few commercial tests based on molecular strategies available for the detection of these pathogens. One of these tests is the Oligo Speed here compared to conventional PCR-based methods targeting different genes for each microbial agent. A great concern on this matter lately raised is the intra-strain gene polymorphism found within meningococcal strains as described on a study.20 False-negative results by real-time PCR were described for the ctrA gene used as a diagnostic target. Our results show that the combination of specific primers targeting genes nspA, ply, and P6 on a multiplex approach was able to detect at least 10 gene copies of each bacterial agent.
Two variants of H. influenzae causing either invasive or non-invasive disease have been described. The most common is the capsulated Hi variant whose capsule is classified according to its polysaccharide structure into six different serotypes (a, b, c, d, e and f) which can be detected using PCR.19 However, this approach fails to identify non-capsulated strains since it is based on the amplification of a genetic capsule structure. The most common virulence factor of Hi strains is the capsule which helps the bacteria to evade the host immune system. Nevertheless, an increasing number of non-typeable strains has been reported causing different forms of disease21–23 and the occurrence of these strains associated to invasive and non-invasive disease has been reported as a concern for new vaccines design.24,25 van Ketel et al.18 described an outer membrane protein (P6) which is exclusively found in all H. influenzae strains and it is not related to the presence of the capsule, being suitable to discriminate between capsulated and non-capsulated strains through a simple PCR. Failure to amplify the bexA gene and amplification of the P6 gene indicates a non-capsulated Hi strain. The single use of primers for the detection of the P6 gene would be sufficient for the detection of Hi strains, but this approach alone would not indicate if the strain is non-capsulated, an important information for epidemiologic surveillance after the introduction of the conjugated Hib vaccine. An additional PCR reaction is required targeting the bexA gene to determine if the strain is capsulated and in this case a serotype-specific PCR can be performed for serotype determination as described by Falla et al.19 The inclusion of an additional PCR with primers targeting the Hi capsular gene bexA is important, since it can differentiate capsulated to non-capsulated Hi strains (NTHi) which has been considered as a potential emerging pathogen in countries where the conjugated vaccine against Hib has been introduced.21,24–29
An additional advantage of the multiplex system here described is that there is no possibility of misinterpretation of the specific bands with primer excess, which is frequently found below 100bp, since the length of the shortest band is 273bp for the bexA gene fragment and other bands are located at 343bp (P6), 481bp (nspA) and 1000bp (ply). Other PCR-based methods use real-time PCR primers which may generate short-length amplicons with difficult preview by agarose gel electrophoresis.7,10
Sensitivity was assessed using clinical samples (CSF and whole blood) from culture-confirmed cases for one of the three organisms. Sensitivity for Nm, Sp, and Hi was 100% since all positive cases confirmed by culture gave positive PCR result. Furthermore, 12 samples associated to suspected cases were also scored positive by the multiplex PCR approach here described. It is likely that these samples were considered negative due to previous antibiotic treatment. The use of the novel multiplex PCR gave a total of 46 (34.3%) PCR positive samples (Table 3). These results showed how sensitive is the test when other phenotypic or epidemiological results indicate that the agent causing disease could be one of those three bacterial pathogens. Problems related to culture and isolation of these bacteria are frequently reported by several laboratories, where a specific and sensitive molecular test is crucial for the rapid and accurate identification of the disease causing agent.
Real-time PCR has been pointed as advantageous over conventional PCR due to its higher sensitivity and faster resolution since the result can be monitored during the run. However, real-time PCR does not appear to offer advantages when used by public laboratories for broad-range PCR testing a large number of samples. In fact, there may be disadvantages, such as need of trained technical staff, controlled environment for assay setups, and higher cost of equipment and reagents. For public or private laboratories that still use conventional PCR to detect such pathogens, the choice for the best multiplex system is crucial to achieve accurate and reliable results. In the present study, we were able to compare several different primers combinations and gene targets recently described by different authors, with a commercial PCR-based test for the detection of the three agents. Our data show that the most sensitive and reliable combination of primers for a multiplex approach include the following targets: nspA gene for N. meningitidis generating a 481bp amplicon,8ply gene for S. pneumoniae generating a 1000bp amplicon (sequence described in this study), and P6 gene for H. influenzae generating a 273bp amplicon.18 For the discrimination between capsulated and non-capsulated strains of H. influenzae, an additional PCR is required, targeting the bexA gene generating a 343bp amplicon,19 when a positive amplification of the P6 gene is observed.
We believe that the standardization of conventional PCR-based tests for the detection of the invasive bacterial agents here described could improve, interoperability, reproducibility, and quality among different worldwide laboratories involved in routine clinical analysis.
FundingFinancial support was provided by INCQS/FIOCRUZ and FAPERJ Proc.N.E26/110.761/2010.
Conflicts of interestThe authors declare no conflicts of interest.
The authors are grateful to the Sequencing core “Plataforma Genomica de Sequenciamento de DNA/PDTIS-FIOCRUZ”. The study was approved by the local Ethics Committee.