VanA glycopeptide resistance is characterized by acquired inducible resistance to both vancomycin and teicoplanin, whereas the VanB phenotype is characterized by variable levels of resistance to vancomycin but susceptibility to teicoplanin.1 Here we report a vancomycin-resistant Enterococcus faecium (VREFm) strain with VanB phenotype-vanA genotype incongruence.
In the course of antimicrobial susceptibility testing of all Enterococcus spp. isolates, we identified a VREFm strain (BG139/2013) exhibiting high-level resistance to vancomycin (MIC>256μg/mL) and susceptibility to teicoplanin (4μg/mL) according to the Clinical and Laboratory Standards Institute 2013 recommendations, hence having the VanB phenotype. BG139/2013 strain was isolated on 22 February 2013 from the blood of a male patient aged 37 years at a Department of Nephrology and Dialysis in Gabrovo, Bulgaria. Species identification was done by the VITEK 2 system (bioMérieux) and confirmed using multiplex polymerase chain reaction (PCR) as previously described.2
Antimicrobial susceptibility testing was performed by the Etest (LIOFILCHEM). BG139/2013 showed high-level gentamicin resistance; resistance toward ampicillin (MIC>256μg/mL), erythromycin (>256), tetracycline (24), ciprofloxacin (>32); and susceptibility to linezolid (1.5) and chloramphenicol (1.5).
Bacterial DNA was extracted using the ISOLATE Genomic DNA Mini Kit (Bioline, UK) according to the manufacturer's instructions. The glycopeptide antibiotic resistance genes were amplified with specific primers as described before: A1/A2, B1/B23 and vanD-F/vanD-R.4 The primers for the vanA and vanD genes were actualized: vanAD-F 5′-GARGAYGGMWSCATMCARGGY-3′ and vanAD-R 5′-MGTRAAWCCNGGCAKRGTRTT-3′. Each 25-μL PCR mixture consisted of 3μL of template DNA; a 0.1μM of each primer; 12.5μL of MyTaq PCR mix (Bioline) and 7.5μL of ultrapure 18.2MΩ PCR water (Bioline). DNA amplification was performed using the following protocol: initial denaturation (95°C for 5min), followed by 35 cycles of denaturation (95°C for 45s), annealing (51.3–60°C for 45s) and extension (72°C for 60s), with a single final extension of 7min at 72°C. For primer modification and phylogenetic analyses we used the GenBank NCBI (National Center for Biotechnology Information) sequence database: FJ545640, JN207933, JN207928, JN207930, EF206284, X56895 (VanA strains); AY665551, U35369, AF310956 and AY145441 (VanB); AF175293, AF153050, AF130997, AY489045, AY082011 and AF277571 (VanD). The sequence reactions and phylogenetic analyses were performed as previously described.5
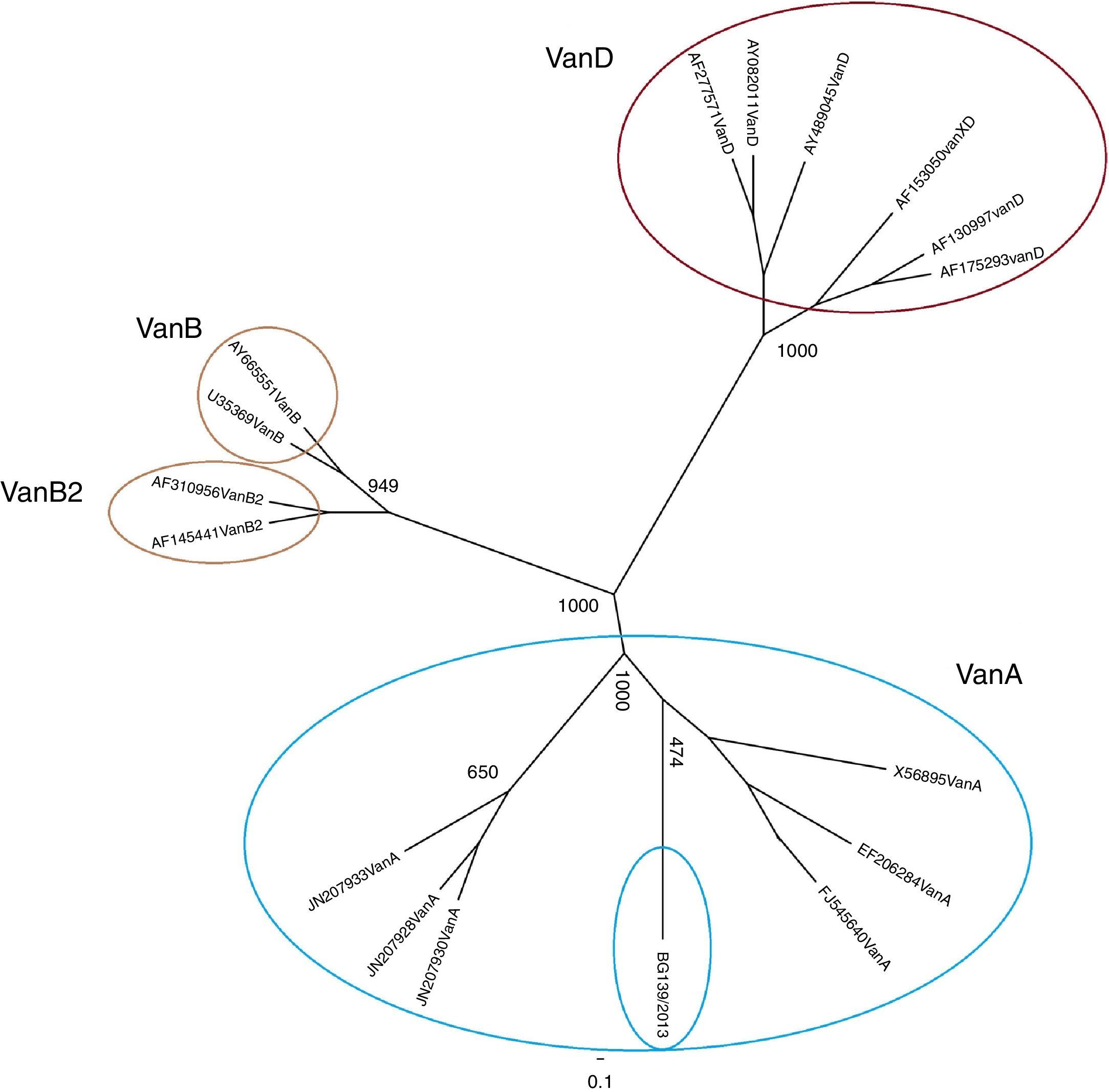
A 630bp PCR product was yielded only with the vanAD-F/vanAD-R primer pair at an annealing temperature of 51.3°C. After sequencing, the data for the phylogenetic analysis consisted of a 450 nucleotides fragment, or 150 amino acid (aa) residues, including 125–274 aa of vanA. The predicted aa sequence of the BG139/2013 isolate was 100% identical to the known vanA sequences (GenBank accession no AAW56079.1 and AGU36307.1). The alignment with JN207933, JN207928 and JN207930 showed that our isolate had a non-synonymous nucleotide mutation leading to the substitution of alanine with valine in position 227 in the amino acid chain. The phylogenetic tree showed that BG139/2013 isolate groups together with cluster vanA gene isolates of E. faecium (Fig. 1).
Phylogenetic tree based on alignment of a 150 amino acid predicted sequence from isolate BG139/2013 with E. faecium reference strains. The tree was constructed from aligned amino acid positions in the final data set using MUSCLE. One thousand bootstrap replications were used to build the phylogenetic tree.
The VanA phenotype resistance is determined by genes located on transposon Tn1546, which includes the vanRS regulatory system that activates the transcription of resistance genes (vanHAXYZ).6 The studied gene fragment makes up a part of the protein active site and was identified as vanA genotype in the phylogenetic analysis. On the other hand, BG139/2013 had VanB phenotype. A relation has been reported between an impairment of the response to teicoplanin (i.e. low-level teicoplanin resistance) and amino acid substitutions due to three point mutations in the N-terminal region of vanS.6 Thus, the substitution found in position 227 seems to be unrelated to the discrepancy between the phenotype and genotype. This discrepancy could most probably be attributed to non-synonymous mutations in the N-terminal region of vanS.
In conclusion, further studies on the genes responsible for the phenotype resistance of the BG139/2013 isolate, and on vanS in particular, are needed. To our knowledge, the studied VanB phenotype-vanA genotype E. faecium strain is the first of its kind to be isolated in Bulgaria.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by a grant from the Medical University of Sofia, Bulgaria (Council of Medical Science, project no. 12/2013, grant no. 16/2013).