The human hepatitis B virus causes acute and chronic hepatitis and is considered one of the most serious human health issues by the World Health Organization, causing thousands of deaths per year. There are similar viruses belonging to the Hepadnaviridae family that infect non-human primates and other mammals as well as some birds. The majority of non-human primate virus isolates were phylogenetically close to the human hepatitis B virus, but like the human genotypes, the origins of these viruses remain controversial. However, there is a possibility that human hepatitis B virus originated in primates. Knowing whether these viruses might be common to humans and primates is crucial in order to reduce the risk to humans.
ObjectiveTo review the existing knowledge about the evolutionary origins of viruses of the Hepadnaviridae family in primates.
MethodsThis review was done by reading several articles that provide information about the Hepadnaviridae virus family in non-human primates and humans and the possible origins and evolution of these viruses.
ResultsThe evolutionary origin of viruses of the Hepadnaviridae family in primates has been dated back to several thousand years; however, recent analyses of genomic fossils of avihepadnaviruses integrated into the genomes of several avian species have suggested a much older origin of this genus.
ConclusionSome hypotheses about the evolutionary origins of human hepatitis B virus have been debated since the ‘90s. One theory suggested a New World origin because of the phylogenetic co-segregation between some New World human hepatitis B virus genotypes F and H and woolly monkey human hepatitis B virus in basal sister-relationship to the Old World non-human primates and human hepatitis B virus variants. Another theory suggests an Old World origin of human hepatitis B virus, and that it would have been spread following prehistoric human migrations over 100,000 years ago. A third theory suggests a co-speciation of human hepatitis B virus in non-human primate hosts because of the proximity between the phylogeny of Old and New World non-human primate and their human hepatitis B virus variants. The importance of further research, related to the subject in South American wild fauna, is paramount and highly relevant for understanding the origin of human hepatitis B virus.
The human hepatitis B virus (HBV) is a small enveloped DNA virus that causes two forms of hepatitis – acute and chronic. The World Health Organization (WHO) estimates that more than 1/3 of the world population already possess serologic evidence of HBV infection, 350 million people are chronically infected and approximately 600,000 deaths are reported each year as a result of acute or chronic hepatitis B.1,2 Infected patients have a high risk of developing serious problems related to the disease, such as fibrosis, cirrhosis and hepatocellular carcinoma (HCC).3 Today, HCC is the fifth most frequent human cancer.4,5
A prophylactic vaccine against a subunit of the hepatitis B surface antigen (HBs) is already available. However, there can be failures in some cases such as emerging anti-HBs-escape mutant viruses, patients with compromised immune-response or heterologous HBV genotypes.6–8 Despite the safety and efficacy of the developed vaccines, infection with hepatitis B virus (HBV) continues to persist as a public health problem and a leading cause of death from infectious diseases worldwide. HBV can be divided into at least ten genotypes (A–J), distinguished by more than an 8% difference in their genomic nucleotide sequence. There are variations in the geographical distribution of HBV genotypes. While the genotypes A, D and G are detected all over the world, genotypes B, C, I and J are most common in Asia, genotype F and H in Native Americans, and genotype E can be found in Western Africa.9–11
The Hepadnaviridae family is made up of genera Orthohepadnavirus and Avihepadnavirus. The former infects mammals and is represented by: HBV (Hepatitis B Virus), which infects humans; viruses similar to HBV (such as woolly monkey HBV and orangutan-HBV for example), which infect non-human primates; Woodchuck hepatitis virus (WHV) that causes hepatitis in woodchucks, and Ground squirrel hepatitis virus (GSHV) causing hepatitis in squirrels.12,13 The latter infect birds, including Duck hepatitis B virus (DHBV) responsible for duck hepatitis and Heron hepatitis B virus (HHBV) which causes hepatitis in herons.14–16
General characteristics of Hepadnaviridae familyHepadnaviridae family members have as common characteristics a tropism for liver cells. They also possess enveloped virions and icosahedral nucleocapsid; the genome consists of an incomplete double stranded DNA with their own DNA polymerase, which is a long negative strand and a short positive strand, with the latter of a variable length, depending on each species. They produce subviral particles; generate persistent infection and replicate through RNA intermediate via reverse transcriptase.17
The genome of human HBV (which is the prototype species of the family Hepadnaviridae) has a peculiar genomic organization with a mechanism of asymmetric replication. This genome is relaxed circular and has a length of approximately 3200 base pairs and has four regions of open reading frame[s?] (ORF) which overlap: Pré-S1/Pré-S2/S, Pre C/C, X and P. These genes are responsible for encoding the three envelope (or surface) proteins; small (S), medium (M), and large (L), that constitute the HBV surface antigen (HBsAg), the core protein (HBcAg), the nuclear protein (HBeAg), X protein (that can affect viral replication and proliferation, and interfere in cellular processes of apoptosis and carcinogenesis) and viral polymerase (P).18,19 The ORF of the P gene occupies 80% of the entire genome and encodes the viral DNA polymerase enzyme which has three areas: DNA polymerase, reverse transcriptase and RNAase. To better describe mutations in this gene, the reverse transcriptase domain was sub-divided into 7 sub-domains, classified A–G.17,18,20 Mutations in the P gene cause a large increase in the synthesis of viral DNA and greater risk of liver carcinogenesis.18
Another characteristic is that hepatitis B is a non-retroviral virus that uses reverse transcriptase as part of its replication process, using the ‘covalently closed circular DNA’ (cccDNA), which serves as a template for transcription of four viral mRNAs and allows the maintenance of chronic HBV infection in hosts.21
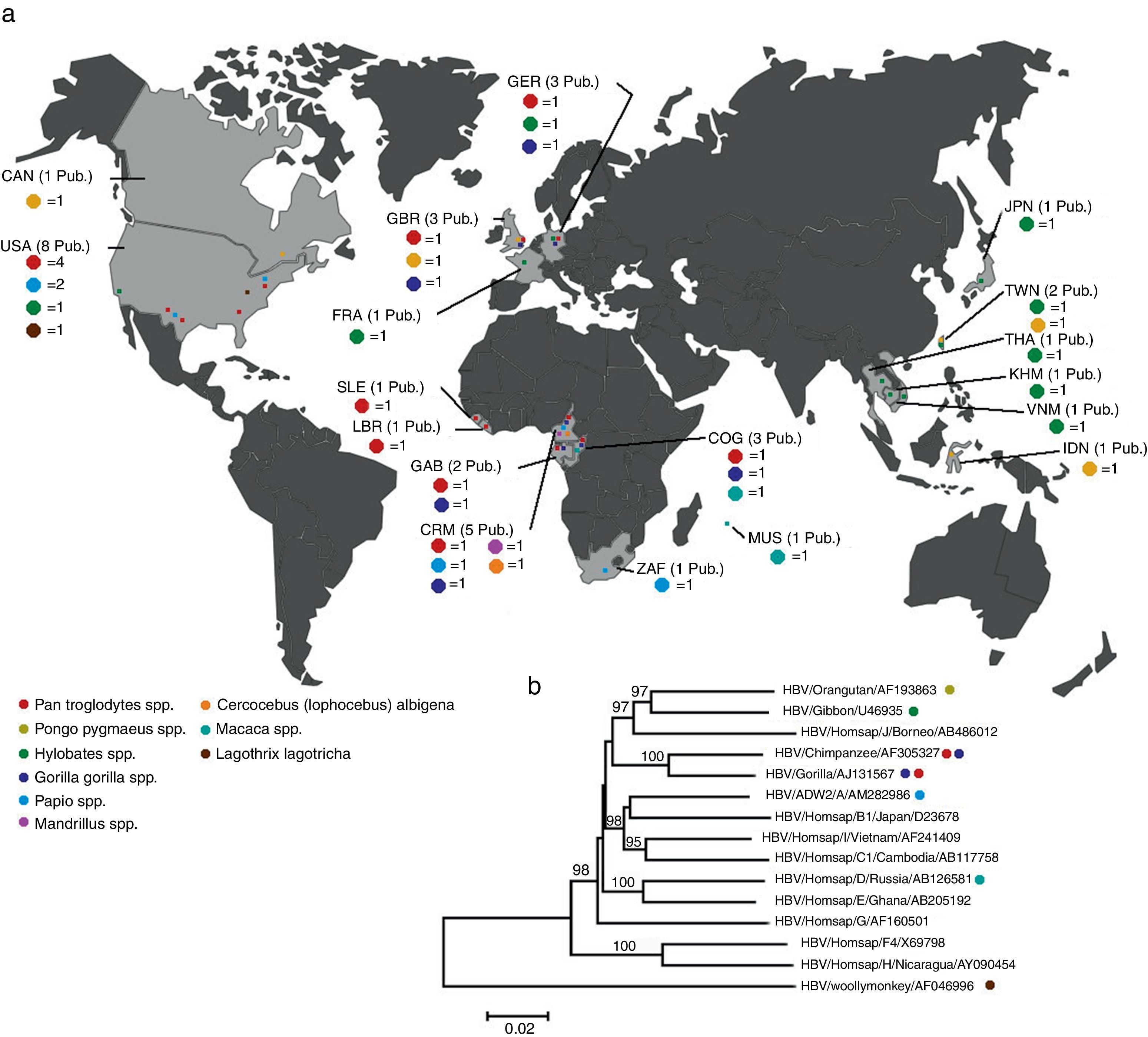
HBV in primatesBesides human HBV, which is the model species of the family, there are viruses belonging to the same family that infect other primates (Table 1). Non-human HBV genotypes were found in higher Old World primates.22,23 Together with the high diversity of African HBV subgenotypes and recombinants, this led to the possibility that the origin of HBV occurred in Old World primates.23,24 Most isolates from non-human primates (NHP) were phylogenetically close to human HBV isolates. Viruses were found in gibbons (Nomascus sp. and Hylobates sp.),25 chimpanzees (Pan troglodytes),26–29 gorillas (Gorilla gorilla)30 and orangutans (Pongo pygmaeus)31 (Fig. 1).
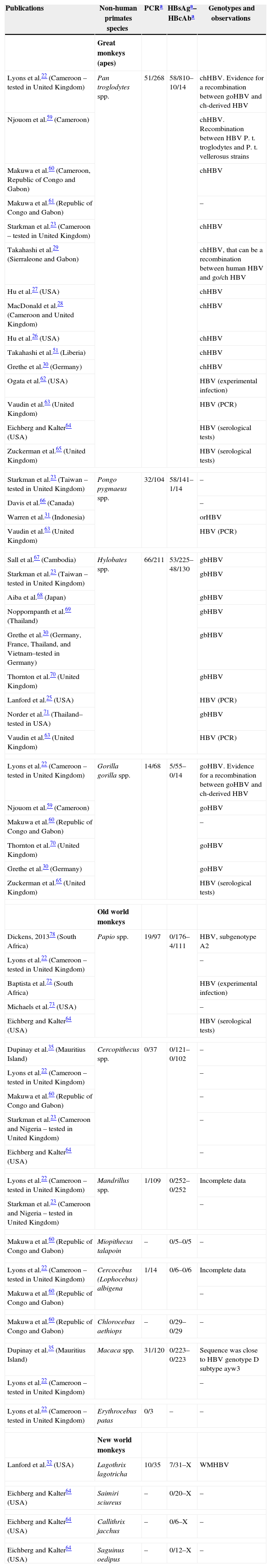
Characteristics of previous publications about HBV infections in primates: PCR detection, HBsAg, HBcAb, genotypes and observations.
| Publications | Non-human primates species | PCRa | HBsAga–HBcAba | Genotypes and observations |
|---|---|---|---|---|
| Great monkeys (apes) | ||||
| Lyons et al.22 (Cameroon – tested in United Kingdom) | Pan troglodytes spp. | 51/268 | 58/810–10/14 | chHBV. Evidence for a recombination between goHBV and ch-derived HBV |
| Njouom et al.59 (Cameroon) | chHBV. Recombination between HBV P. t. troglodytes and P. t. vellerosus strains | |||
| Makuwa et al.60 (Cameroon, Republic of Congo and Gabon) | chHBV | |||
| Makuwa et al.61 (Republic of Congo and Gabon) | – | |||
| Starkman et al.23 (Cameroon – tested in United Kingdom) | chHBV | |||
| Takahashi et al.29 (Sierraleone and Gabon) | chHBV, that can be a recombination between human HBV and go/ch HBV | |||
| Hu et al.27 (USA) | chHBV | |||
| MacDonald et al.28 (Cameroon and United Kingdom) | chHBV | |||
| Hu et al.26 (USA) | chHBV | |||
| Takahashi et al.51 (Liberia) | chHBV | |||
| Grethe et al.30 (Germany) | chHBV | |||
| Ogata et al.62 (USA) | HBV (experimental infection) | |||
| Vaudin et al.63 (United Kingdom) | HBV (PCR) | |||
| Eichberg and Kalter64 (USA) | HBV (serological tests) | |||
| Zuckerman et al.65 (United Kingdom) | HBV (serological tests) | |||
| Starkman et al.23 (Taiwan – tested in United Kingdom) | Pongo pygmaeus spp. | 32/104 | 58/141–1/14 | – |
| Davis et al.66 (Canada) | – | |||
| Warren et al.31 (Indonesia) | orHBV | |||
| Vaudin et al.63 (United Kingdom) | HBV (PCR) | |||
| Sall et al.67 (Cambodia) | Hylobates spp. | 66/211 | 53/225–48/130 | gbHBV |
| Starkman et al.23 (Taiwan – tested in United Kingdom) | gbHBV | |||
| Aiba et al.68 (Japan) | gbHBV | |||
| Noppornpanth et al.69 (Thailand) | gbHBV | |||
| Grethe et al.30 (Germany, France, Thailand, and Vietnam–tested in Germany) | gbHBV | |||
| Thornton et al.70 (United Kingdom) | gbHBV | |||
| Lanford et al.25 (USA) | HBV (PCR) | |||
| Norder et al.71 (Thailand–tested in USA) | gbHBV | |||
| Vaudin et al.63 (United Kingdom) | HBV (PCR) | |||
| Lyons et al.22 (Cameroon – tested in United Kingdom) | Gorilla gorilla spp. | 14/68 | 5/55–0/14 | goHBV. Evidence for a recombination between goHBV and ch-derived HBV |
| Njouom et al.59 (Cameroon) | goHBV | |||
| Makuwa et al.60 (Republic of Congo and Gabon) | – | |||
| Thornton et al.70 (United Kingdom) | goHBV | |||
| Grethe et al.30 (Germany) | goHBV | |||
| Zuckerman et al.65 (United Kingdom) | HBV (serological tests) | |||
| Old world monkeys | ||||
| Dickens, 201378 (South Africa) | Papio spp. | 19/97 | 0/176–4/111 | HBV, subgenotype A2 |
| Lyons et al.22 (Cameroon – tested in United Kingdom) | – | |||
| Baptista et al.72 (South Africa) | HBV (experimental infection) | |||
| Michaels et al.73 (USA) | – | |||
| Eichberg and Kalter64 (USA) | HBV (serological tests) | |||
| Dupinay et al.35 (Mauritius Island) | Cercopithecus spp. | 0/37 | 0/121–0/102 | – |
| Lyons et al.22 (Cameroon – tested in United Kingdom) | – | |||
| Makuwa et al.60 (Republic of Congo and Gabon) | – | |||
| Starkman et al.23 (Cameroon and Nigeria – tested in United Kingdom) | – | |||
| Eichberg and Kalter64 (USA) | – | |||
| Lyons et al.22 (Cameroon – tested in United Kingdom) | Mandrillus spp. | 1/109 | 0/252–0/252 | Incomplete data |
| Starkman et al.23 (Cameroon and Nigeria – tested in United Kingdom) | – | |||
| Makuwa et al.60 (Republic of Congo and Gabon) | Miopithecus talapoin | – | 0/5–0/5 | – |
| Lyons et al.22 (Cameroon – tested in United Kingdom) | Cercocebus (Lophocebus) albigena | 1/14 | 0/6–0/6 | Incomplete data |
| Makuwa et al.60 (Republic of Congo and Gabon) | – | |||
| Makuwa et al.60 (Republic of Congo and Gabon) | Chlorocebus aethiops | – | 0/29–0/29 | – |
| Dupinay et al.35 (Mauritius Island) | Macaca spp. | 31/120 | 0/223–0/223 | Sequence was close to HBV genotype D subtype ayw3 |
| Lyons et al.22 (Cameroon – tested in United Kingdom) | – | |||
| Lyons et al.22 (Cameroon – tested in United Kingdom) | Erythrocebus patas | 0/3 | – | – |
| New world monkeys | ||||
| Lanford et al.32 (USA) | Lagothrix lagotricha | 10/35 | 7/31–X | WMHBV |
| Eichberg and Kalter64 (USA) | Saimiri sciureus | – | 0/20–X | – |
| Eichberg and Kalter64 (USA) | Callithrix jacchus | – | 0/6–X | – |
| Eichberg and Kalter64 (USA) | Saguinus oedipus | – | 0/12–X | – |
(A) Geographical distribution of publication relating to non-human primates which were detected with some HBV genotype. Sample animals are listed by genera in Table 1. (B) The evolutionary history was inferred by Neighbor–Joining method.74 The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown above the branches.75 The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method76 and are in the units of number of base difference per site. All positions containing gaps and missing data were eliminated. There were a total of 3167 positions in the final dataset. evolutionary analyses were conducted in MEGAS.77
The origin of these viruses remains controversial. Chimpanzees from different regions of the planet appear to be infected with different types of isolates,27 supporting the concept that these viruses may have evolved with their hosts. However, the hepadnavirus isolated from woolly monkeys (Lagothrix lagotricha) – woolly monkey HBV (WMHBV) clearly is a new member of the Hepadnaviridae family.32 Both the WMHBV and the New World human HBV genotypes F and H were in a phylogenetic sister-relationship which is more divergent to the Old World human HBV genotypes, strengthening hypotheses on an origin of HBV in New World primates.33,34 Until the present moment, WMHBV has been detected only in captive animals at one zoo, but it was not found in another four zoos tested.32 So far, there have been no reports about the presence of this virus in wild woolly monkeys.
In 2013, Dupinay et al.35 investigated HBV infection in small primates of different origins and in Mauritius, 25.8% (in serum) and 42% (in liver) of HBV DNA positives were found. Genome sequencing data revealed that it was human HBV genotype D, subtype ayw3. This provides evidence for the existence of small model simian chronic HBV infection, immunologically near human and that it might have been transmitted from man hundred years ago.
Theories of HBV origins and evolutionWith the advent of the new field of paleovirology, studies have revealed that integration into the nuclear genome of germline cells can lead to vertical inheritance of retroviral genes as host alleles. Such integration events are referred to as endogenous viral elements (EVEs). Paleoviral “fossils” were recently discovered from a wide range of viruses including EVE non-retroviral families (rtDNA viruses) e.g.: Hepadnaviridae.36,37 Genomic “fossils” of avihepadnaviruses integrated into the genomes of diverse avian species suggest that the origin of this group is much older dating back several million years to the Mesozoic Era.38–40 In a recent study, Suh et al.40 emphasized that “insights into long-term sequence evolution, genome evolution and hypothetical ancestral hosts change the understanding of the prehistoric evolution of Hepadnaviridae, including the hypothetical origin of mammalian HBVs,” contributing to several studies in the field of medical and paleontological research. The evolutionary origin of hepadnavirus in primates has been dated back several thousand years.33 No “fossil” records have been described from primates so far.
Most of our knowledge about pathogenesis and the replication of hepatitis B virus has been obtained from studies of hepadnaviruses related to animals as models, including Duck Hepatitis B Virus (DHBV),41 Woodchuck Hepatitis virus (WHV),42 Ground Squirrel Hepatitis virus (GSHV),43 Arctic Squirrel Hepatitis virus (ASHV),44 Heron Hepatitis B Virus (HHBV),45 and Stork Hepatitis B Virus (STHBV).46
Studies about pathogens that affect primates are of great scientific relevance because of their evolutionary relationship with humans. Non-human primates can be sentinels for surveillance of infectious agents and also biological models for important diseases affecting the human species, such as HBV. Checking for diseases common to humans and primates is crucial in reducing risks to the health of humans.47
All known hepadnaviruses that affect primates and other animals cause acute and chronic hepatic infection in their hosts.48 A main characteristic of all the viruses in this family is the possibility of chronic infection with serious clinical consequences.48,49 However, what are the implications of the origins of HBV?
During the 1990s, three competing hypotheses about the evolutionary origins of HBV emerged, as reviewed by Simmonds in 2001.24 A New World origin was proposed because of the phylogenetic co-segregation between the New World HBV genotypes F and H and WMHBV in basal sister-relationship, to the Old World non-human primates and human HBV variants. The dissemination of HBV into the Old World would then have occurred either in the post-Columbian Era (nearly 500 years ago) or during earlier human migrations about 15,000 years ago.33,34 A second theory suggests an Old World origin of HBV, and that it was spread following prehistoric human migrations over 100,000 years ago.50 A third theory suggests a co-speciation of HBV in non-human primate hosts because of the proximity between the phylogeny of Old and New World NHP and their HBV variants.28
These theoretical approaches were unable to reconcile conflicting data resultant from the distribution of HBV variants in human and non-human primate hosts on a global scale. An example is that the HBV genotypes in Old World primates are mutually equidistant. This is unlikely to have occurred from independent acquisitions of human HBV variants in only a few hundred years, going against the New World origin theory.24 In relation to the theory of a supposed exodus of HBV together with human migration out of Africa to other areas of the planet does not explain the distribution of HBV genotypes in the world.24 For example, the New World HBV genotype F that is found in Native Americans is also found in genetically unrelated island populations of Polynesia. The closest relatives of Native Americans living in Northeastern Asia are predominantly infected by genotypes B and C of HBV.24
The widely host-specific HBV variants support the theory about HBV co-speciation with primate hosts. However, as noted by Simmonds,24 this theory is challenged by a lack of shared HBV variants between humans and non-human primates, by the existence of cross-species HBV transmission and by the lack of higher diversification in NHP compared to human HBV variants. These have long been deemed unlikely, but recent data has shown that orangutans and gibbons sharing habitats in Asia harbour phylogenetically co-segregating HBV variants. Similarly, recombination events between HBV genotypes of chimpanzee and gorilla from Central Africa, phylogenetically grouped together and inter-species have been described.22,23 Furthermore, sporadic detections of human-related HBV genotypes have been described in chimpanzees and Orangutans, both of the Hominidae family.9,51 Cui52 suggests that a large number of cross-species transmission events are probably also present in avihepadnaviruses, whose phylogeny does not match that of the host orders Gruiformes and Anseriformes.52 Gibbons separated genetically from orangutans, gorillas and chimpanzees about 20 million years ago. Despite this, the orangutan and gibbon HBV variants found to date are similar.22
Evidence of animals as reservoirs, possible recombination between human and non-human primate HBV variants, and inter-species transmissions all provide important information that may assist in the eradication of HBV throughout the world.28
Co-evolution of HBV variants in non-human primates is further challenged by the comparable degree of diversification within viruses that affect chimpanzees, gibbons and gorillas, and phylogenetic sister-relationships between these viruses that do not reflect the evolution of their hosts. This includes the absence of outlier gorilla HBV variants in relation to chimpanzee variants and human HBV genotypes F and H in phylogenetic outlier position to the Old World NHP viruses.22 Similarly, this theory fails to explain the position of gibbon HBV variants and their comparable diversification to other primate HBV genotypes (phylogenetically), while they should be more diversified than those of great apes given their divergence from the latter about twenty million years ago.52 Co-evolution on the scale of the Hepadnaviridae family seems even less likely, given the high diversity of rodent and bird hepadnaviruses from their primate counterparts. As noted above, this does not match the timescale of the likely separation of these variants, which may be only 10–20 times greater than the development of monkey.22
Attempts to determine a non-recent Hepadnaviridae evolution are compromised by several factors. This includes the sequence constraints arising from largely overlapping ORFs.33 While this technical problem can be dealt with,33 there are several recent descriptions of avihepadnavirus genomic fossils.38,39,53 These sequences have indicated that previous assumptions based on existent HBV variants may have underestimated the period of hepadnaviruses origin in the range of several million years.38 Theoretically, a continuous appearance of infectious viruses from genomic pockets could explain the unusually slow rates of sequence change in the range of 10 per site per year observed that are necessary to explain the sequence divergence between non-human primates HBV hosts in a co-speciation scenario.24,33
Understanding the relationship between humans and NHP variants of HBV will help explain the possible role that humans and other primates have played in the spread of HBV around the globe.22 No hepadnavirus elements in primate genomic data have been described to date. It is clear that one single theory has been unable to explain the emergence of HBV until now.
Primates in South AmericaThe most plausible scenario to explain the arrival of primates in South America consists of a transatlantic migration at the end of the Eocene epoch. South American platyrrhine primates probably dispersed from either Asian or African catarrhine ancestors about 37–40 million years ago. However, the radiation of platyrrhines in the New World probably occurred during the Miocene epoch,52,54 although the feasibility of this route is the subject of several debates. The oldest primate fossil discovered in South America is dated to about 27 million years ago.50 Whether Old World hepadnaviruses could have been taken to today's South America by the natural barriers formed by the separation of the sub-continent from the Gondwanan supercontinent about 100 million years ago28 is something that remains unknown. Hypothetical viruses would then have been passed to several host lineages, because a South American evolution of platyrrhines 100 million years ago is deemed highly unlikely.28
The high degree of complexity in epidemiological factors such as transmission routes can affect the large number of HBV variants in circulation. However, the propagation of these viruses can be enhanced with the development of recombinant variants, allowing the virus to be transmitted more efficiently between different species.10,55
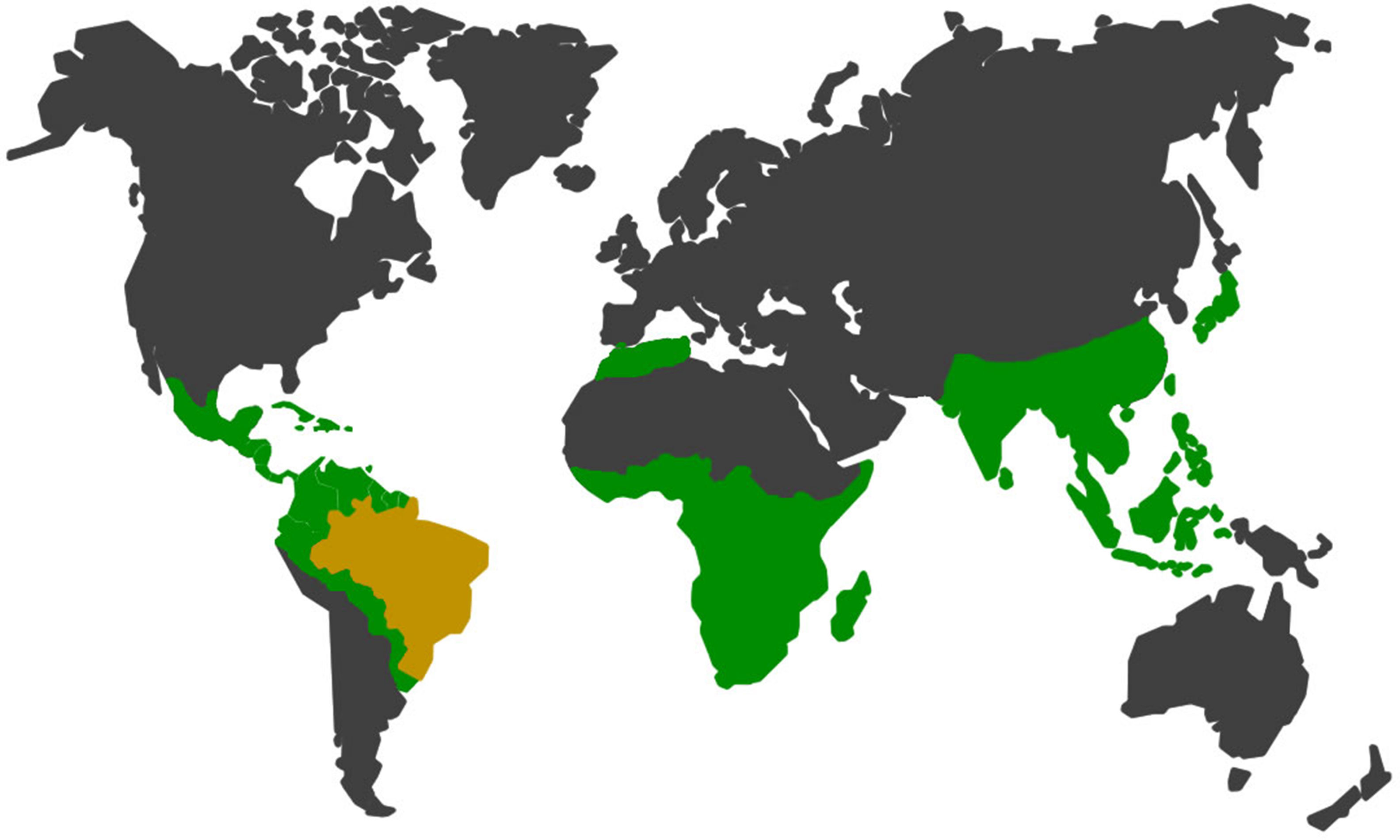
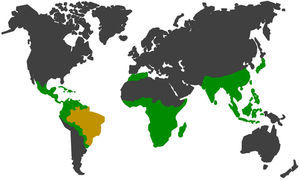
There are many primates in South America, and in Brazil alone 133 species and subspecies of Neotropical primates can be found in the wild fauna, which represents approximately 21% of the existing group on the planet56 (Fig. 2). Wild cycles and urban cycles of transmission of some pathogens start to intertwine forming new epidemiological settings with risks to human and animal populations in both environments.57,58
In green and yellow, the geographic distribution of non-human primates52,54,56,57. In yellow, Brazil which has large range of species and subspecies of neotropical non-human primates in the wild fauna, representing approximately 21% of the group in the world.56,57
Until now there has been no study into infectious hepadnavirus in non-human primates of the fauna in South America and it is not known whether the existing species in the New World may or may not be carrying the hepatitis B virus that infects humans, or if it is another variant of the Hepadnaviridae family. Therefore, to investigate the presence of these viruses in this area it is important to try to help unravel the origin of the Hepadnaviruses and how they evolved in primates and humans.
Conflicts of interestThe authors declare no conflicts of interest.