Pseudomonas aeruginosa is an important causative agent of nosocomial infections. As pathogen, P. aeruginosa is of increasing clinical importance due to its ability to develop high-level multidrug resistance (MDR).
MethodsThe aim of the present study was to better understand the intrinsic virulence of circulating strains of Pseudomonas aeruginosa, by surveying and characterizing the antibiotic resistance profiles and prevalence of virulence factors in 51 clinical isolates of P. aeruginosa obtained from children admitted to Hospital del Niño-Panamá during the period of October 2016 until March 2017. Antimicrobial susceptibilities were assessed by determining the minimum inhibitory concentration for 12 antibiotics against P. aeruginosa clinical isolates using the VITEK system (https://www.biomerieux.com). Additionally, all isolates were examined by Polymerase Chain Reaction (PCR) for the presence of components of the MexAB-OprM efflux pump system (mexABR) and pyoverdine receptor genes and betalactamases resistance genes (ESBL) using gene-specific primers.
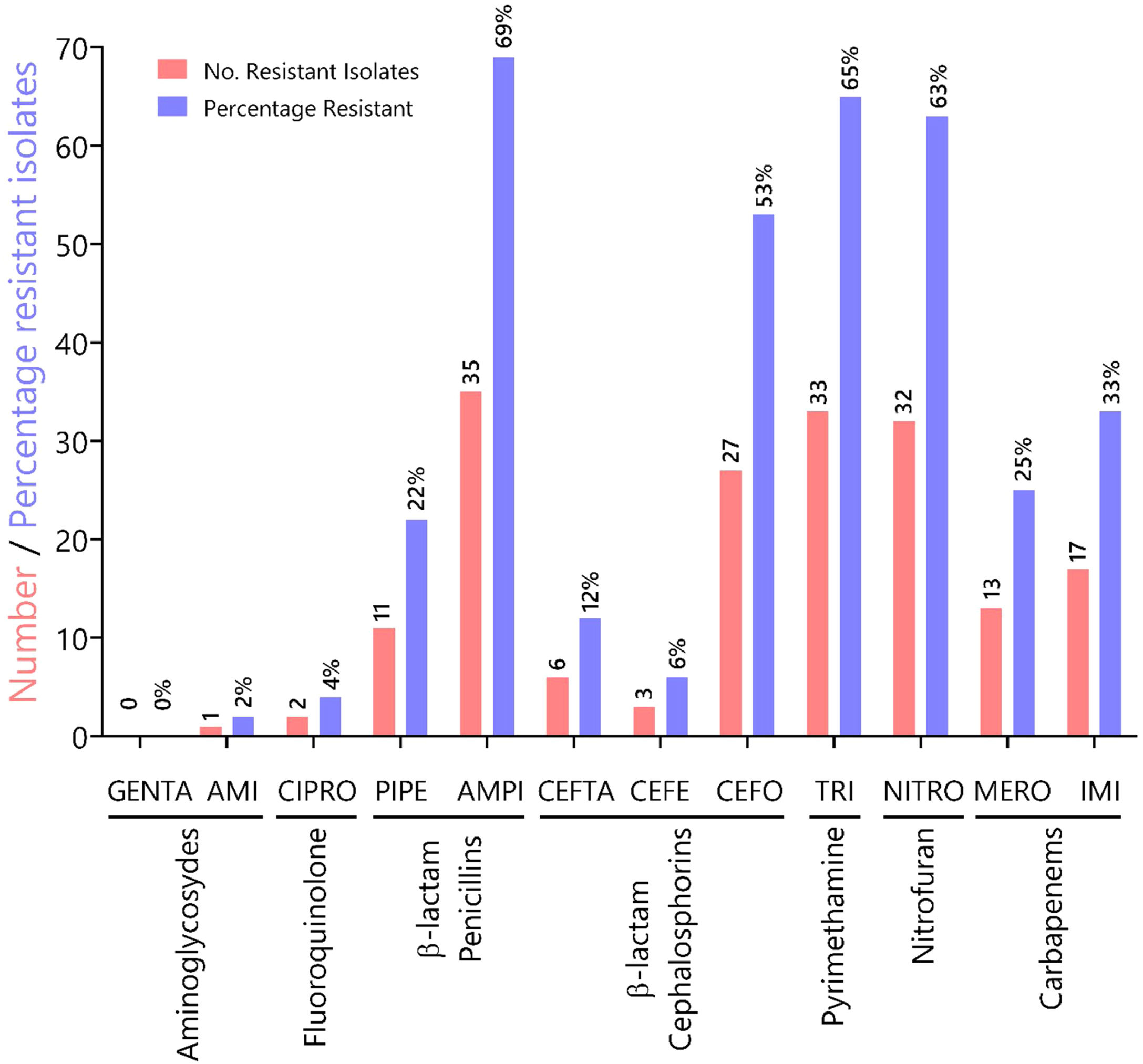
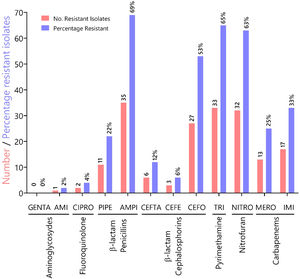
ResultsA total of 51 pyoverdine producing clinical isolates were analyzed, all of which expressed resistance genes such as genes of the MexAB-OprM efflux pump system (mexABR) and pyoverdine receptor genes (fpvA). Out of 51 MDR isolates, 22 were ESBL producers. The most common ESBL gene was blaTEM expressed by 43% of the isolates. The isolates tested in this study showed increased resistance to antibiotics in the following categories: (i) penicillins (ampicillin (69%), piperacillin (22%); (ii) pyrimethamines (trimethoprim, 65%); (iii) nitrofurans (nitrofurantoin, 63%), and (iv) third-generation cephalosporin cefotaxime (53%). These results underscore a high prevalence of MDR amongst clinical isolates from Panama.
ConclusionsThe present study indicates that prevalence of BlaTEM-carrying strains is increasing with subsequent multidrug resistance in Panamá and as well reported worldwide. The virulent factors identified in this study provide valuable information regarding the prevalence of resistance genes and their potential impact on treatments that exploit the unique physiology of the pathogen. To prevent further spread of MDR, the proportions of resistant strains of Pseudomonas aeruginosa should be constantly evaluated on healthcare institutions of Panamá. More importantly, this information can be used to better understand the evolution and dissemination of strains hoping to prevent the development of resistance in Pseudomonas aeruginosa. Future studies quantifying the expression of these virulent genes will emphasize on the acquisition of multidrug resistance.
Pseudomonas aeruginosa is an important causative agent of nosocomial infections. This bacterium is considered an opportunistic pathogen that affects the health of immunocompromised individuals such as those with diabetes, cancer, cystic fibrosis, advanced HIV infections (acquired immunodeficiency syndrome, AIDS), severely burn patients and those that underwent major surgeries.1
As pathogen, Pseudomonas aeruginosa is of increasing clinical importance because of their innate resistance to multiple agents and their ability to develop high-level multidrug resistance (MDR) due to the presence of several virulence factors encoded in its genome (<-- -->Poole, 2016). Several studies have shown that this innate resistance is directly associated to the expression of bacterial efflux pumps and porins such as the Resistance-Nodulation-cell-Division (RND)-type efflux pump, MexAB-OprM, MexC–MexD–OprJ, MexE–MexF–OprN, and MexXY-OprM.2 These important RND-type efflux pumps are constitutively expressed in wild-type cells and are responsible for the intrinsic resistance to most antimicrobial agents.3–7
The MexAB-OprM system was initially characterized from an isolate of P. aeruginosa cultured under iron-depleted conditions in the presence of the iron chelator 2,2’- dipyridyl (Dipy).8,9 Under this condition, protein levels for the MexAB-OprM members were increased, suggesting that the efflux pump plays an essential role in its survival under iron deficiency conditions8,10,11 (Oglesby-Sherrouse et al., 2009)<-- -->.
The MexAB-OprM system includes the proteins MexA, MexB, OprM, and MexR. MexB is the molecular subunit that selectively recognizes specific antibiotics and mediates their expulsion; MexA functions as the interconnecting bridge between OprM and MexB. OprM is a lipoprotein localized in the outer membrane whose role is mediating the final expulsion of the antibiotic across the bacterial outer membrane. MexR acts as repressor of the MexAB-OprM system and autoregulates MexR.12
In addition to the MexAB-OprM efflux pump system, Pseudomonas aeruginosa produce small molecules known as siderophores, which are iron-chelating molecules that contributes to their virulence; siderophores have also been connected to mechanisms of antibiotic resistance (<-- -->Djapgne et al., 2014). Under iron-limiting conditions, P. aeruginosa produces two main siderophores, pyoverdine and pyochelin, which scavenge iron from host proteins, contributing to its virulence.13–15 The ferri-pyoverdine complex uptake is carried out by TonB-dependent receptors with the help of the transporter FpvB,16 resulting in its internalization into the periplasm.17 In addition to its molecular function, the pyoverdine receptor genes have also been used to genotype several strains of P. aeruginosa.18
Another important resistance mechanism in Gram-negative bacteria such as P. aeruginosa is the production of β-lactamases, hydrolytic enzymes involved in the degradation of antibiotics (β-lactams). Among this group, the extended-spectrum β-lactamases (ESBLs) represent an important sub-class of enzymes that confer resistance to oxyimino-cephalosporins, such as cefotaxime (CTX), ceftriaxone, and ceftazidime (CAZ), and the monobactam aztreonam (ATM). Because of this enhanced resistance phenotype against some of the most effective antibiotics, strains carrying ESBLs genes can become a serious clinical problem.19
Several genes coding β-lactamases have been identified at low frequency in P. aeruginosa.20 blaTEM-1 was the first plasmid-encoded β-lactamase gene isolated from E. coli; this isolate came from a blood culture of a patient named Temoniera in Greece, hence the acronym TEM.21 blaTEM-1-like β-lactamases have also been found in other species of the family Enterobacteriaceae and other bacterial species such as Pseudomonas aeruginosa, Haemophilus influenzae, and Neisseria gonorrhoeae.22 Among the ESBL sub-class, SulfHydryl Variable-2 (SHV-2) ESBLs were first identified in a German strain of Klebsiella ozaenae; this novel enzyme is capable of hydrolyzing cefotaxime and to a lesser extent ceftazidime.23 Several other studies have reported the existence of non-blaTEM and non-blaSHV hydrolytic enzymes that do not share the evolutionary and functional origin of typical EBSLs.24 These non-blaTEM and non-blaSHV plasmid-encoded β-lactamases and cefotaximases (blaCTX-M) are able to hydrolyze cephalothin and cefotaxime, and to lesser extend benzylpenicillin and ceftazidime.23 Therefore, the characterization of β-lactamases from drug-resistant P. aeruginosa can greatly contribute to the understanding of mechanisms of molecular pathogenesis, antibiotic resistance and virulence in bacteria<-- -->.25,26
To better understand the intrinsic virulence of circulating strains of Pseudomonas aeruginosa from Panamá, we surveyed and characterized the antibiotic resistance profiles and the prevalence of virulence factors in 51 clinical isolates of P. aeruginosa obtained from children admitted to Hospital del Niño-Panamá.
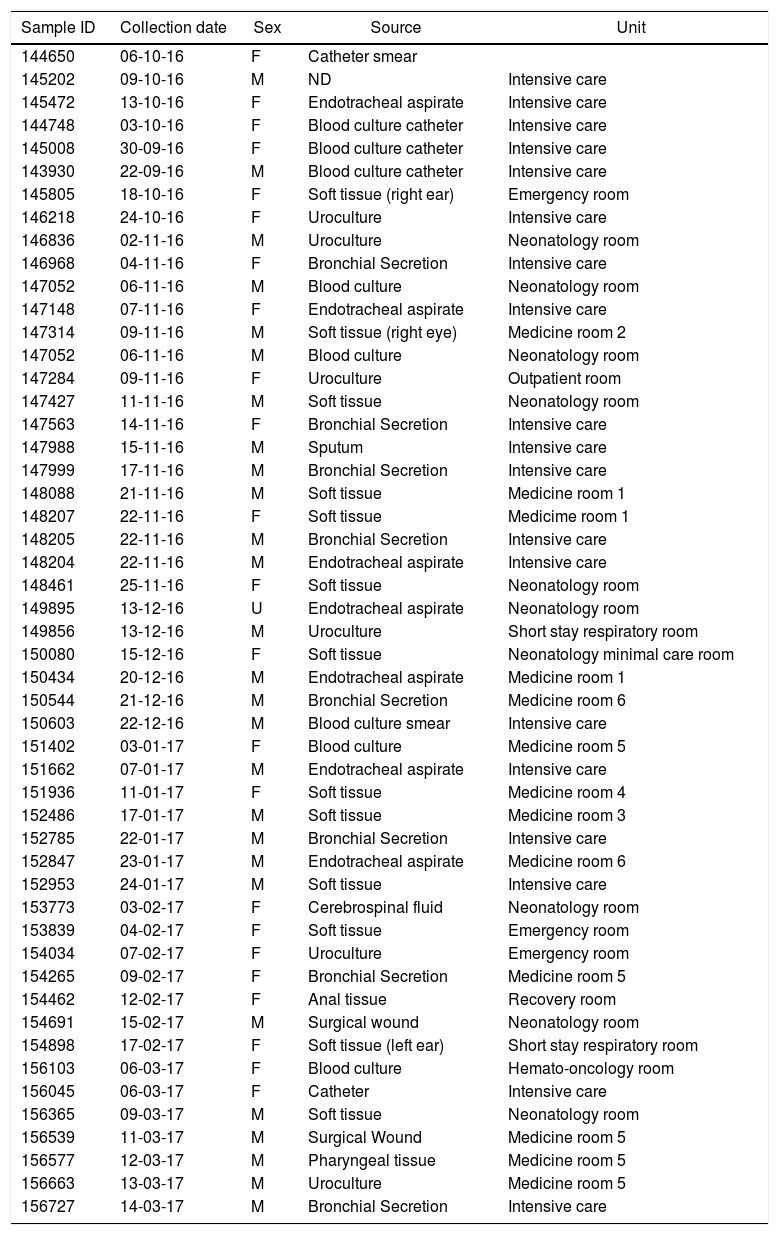
Materials and methodsBacterial Isolates and culture methodsFifty-one strains of P. aeruginosa were isolated from various sites of infections (blood, tracheal secretions, wound, eye and ear discharge, and ulcer) from patients admitted to Hospital del Niño, a Children’s Hospital located in Panama City, Panamá (Table 1). These biological samples were collected by the Hospital’s Microbiology Laboratory between October 2016 and March 2017. In brief, bacterial isolates were grown in Amies transport media with charcoal (Thermo Fisher Scientific, United States) for initial pathogen collection and growth and Bacto Casamino Acids medium (CAA) (Becton-Dickinson, United States) to detect pyoverdine secretion (green-fluorescent pigment). For pyoverdine production, samples were incubated at 37 °C for 24 hours as previously described.27 All strains were store in 50% glycerol at -80 °C. The strain P. aeruginosa ATCC 27853 was used as reference for this study.
Date of isolation, source, patient sex and hospital unit where the strains were identified.
| Sample ID | Collection date | Sex | Source | Unit |
|---|---|---|---|---|
| 144650 | 06-10-16 | F | Catheter smear | |
| 145202 | 09-10-16 | M | ND | Intensive care |
| 145472 | 13-10-16 | F | Endotracheal aspirate | Intensive care |
| 144748 | 03-10-16 | F | Blood culture catheter | Intensive care |
| 145008 | 30-09-16 | F | Blood culture catheter | Intensive care |
| 143930 | 22-09-16 | M | Blood culture catheter | Intensive care |
| 145805 | 18-10-16 | F | Soft tissue (right ear) | Emergency room |
| 146218 | 24-10-16 | F | Uroculture | Intensive care |
| 146836 | 02-11-16 | M | Uroculture | Neonatology room |
| 146968 | 04-11-16 | F | Bronchial Secretion | Intensive care |
| 147052 | 06-11-16 | M | Blood culture | Neonatology room |
| 147148 | 07-11-16 | F | Endotracheal aspirate | Intensive care |
| 147314 | 09-11-16 | M | Soft tissue (right eye) | Medicine room 2 |
| 147052 | 06-11-16 | M | Blood culture | Neonatology room |
| 147284 | 09-11-16 | F | Uroculture | Outpatient room |
| 147427 | 11-11-16 | M | Soft tissue | Neonatology room |
| 147563 | 14-11-16 | F | Bronchial Secretion | Intensive care |
| 147988 | 15-11-16 | M | Sputum | Intensive care |
| 147999 | 17-11-16 | M | Bronchial Secretion | Intensive care |
| 148088 | 21-11-16 | M | Soft tissue | Medicine room 1 |
| 148207 | 22-11-16 | F | Soft tissue | Medicime room 1 |
| 148205 | 22-11-16 | M | Bronchial Secretion | Intensive care |
| 148204 | 22-11-16 | M | Endotracheal aspirate | Intensive care |
| 148461 | 25-11-16 | F | Soft tissue | Neonatology room |
| 149895 | 13-12-16 | U | Endotracheal aspirate | Neonatology room |
| 149856 | 13-12-16 | M | Uroculture | Short stay respiratory room |
| 150080 | 15-12-16 | F | Soft tissue | Neonatology minimal care room |
| 150434 | 20-12-16 | M | Endotracheal aspirate | Medicine room 1 |
| 150544 | 21-12-16 | M | Bronchial Secretion | Medicine room 6 |
| 150603 | 22-12-16 | M | Blood culture smear | Intensive care |
| 151402 | 03-01-17 | F | Blood culture | Medicine room 5 |
| 151662 | 07-01-17 | M | Endotracheal aspirate | Intensive care |
| 151936 | 11-01-17 | F | Soft tissue | Medicine room 4 |
| 152486 | 17-01-17 | M | Soft tissue | Medicine room 3 |
| 152785 | 22-01-17 | M | Bronchial Secretion | Intensive care |
| 152847 | 23-01-17 | M | Endotracheal aspirate | Medicine room 6 |
| 152953 | 24-01-17 | M | Soft tissue | Intensive care |
| 153773 | 03-02-17 | F | Cerebrospinal fluid | Neonatology room |
| 153839 | 04-02-17 | F | Soft tissue | Emergency room |
| 154034 | 07-02-17 | F | Uroculture | Emergency room |
| 154265 | 09-02-17 | F | Bronchial Secretion | Medicine room 5 |
| 154462 | 12-02-17 | F | Anal tissue | Recovery room |
| 154691 | 15-02-17 | M | Surgical wound | Neonatology room |
| 154898 | 17-02-17 | F | Soft tissue (left ear) | Short stay respiratory room |
| 156103 | 06-03-17 | F | Blood culture | Hemato-oncology room |
| 156045 | 06-03-17 | F | Catheter | Intensive care |
| 156365 | 09-03-17 | M | Soft tissue | Neonatology room |
| 156539 | 11-03-17 | M | Surgical Wound | Medicine room 5 |
| 156577 | 12-03-17 | M | Pharyngeal tissue | Medicine room 5 |
| 156663 | 13-03-17 | M | Uroculture | Medicine room 5 |
| 156727 | 14-03-17 | M | Bronchial Secretion | Intensive care |
Minimum Inhibitory Concentrations (MICs) of selected antibiotics against P. aeruginosa clinical isolates were evaluated using the VITEK system (bioMérieux) according to manufacturer’s instructions. Susceptibility testing results were based on guidelines set by the European Committee for Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2020_manuals/Manual_v_8.0_EUCAST_Disk_Test_2020.pdf). As main criteria of MDR, a P. aeruginosa isolate was considered as non-susceptible if it displayed resistance to ≥1 antimicrobial agent in ≥3 antimicrobial assays. The antimicrobial agents assayed in this study included (i) aminoglycosides (amikacin - AMI) and gentamicin - GENTA), (ii) fluoroquinolones (ciprofloxacin - CIPRO), (iii) β-lactam-cephalosporins (ceftazidime - CEFTA), cefepime - CEFE), cefotaxime - CEFO), (iv) β-lactam-penicillins (extended spectrum) (piperacillin - PIPE), ampicillin -AMPI)), pyrimethamines (trimethoprim - TRI), (iv) nitrofurans (nitrofurantoin - NITRO) and (v) carbapenems (meropenem - MERO) and imipenem - IMI).
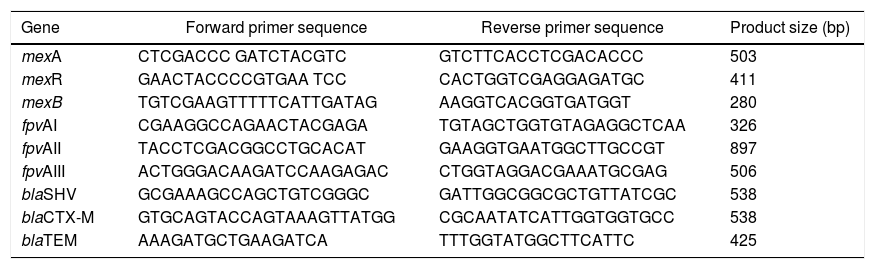
DNA Extraction and Molecular Detection of Resistance GenesIsolates were grown in CAA liquid medium overnight at 37 °C and DNA was extracted from 3 ml of culture following the method for Gram-negative bacteria as described previously (<-- -->Chen and Kuo, 1993). All PCR reactions were prepared using Omega Bio-Tek’s, 2x Taq Master Mix, 0.5 pmol of each primer in 25 μl reactions. The mexABR genes were detected by multiplex PCR using primers listed in Table 2. Cycling conditions consisted of an initial denaturation step at 94 °C for 3 minutes, followed by 32 cycles of 94 °C for 30 seconds, 57 °C for 45 seconds, and 72 °C for 1 minute as described in Auda Al-Grawi et al.12 Pyoverdine receptor genes (fpvA) were amplified simultaneously by multiplex PCR listed in Table 2. The PCR conditions were first a denaturation at 94 °C for 3 min, followed by 30 cycles with denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and elongation at 72 °C for 30 s, and terminating with a last cycle at 72 °C for 10 min.16 To detect the β-lactamase genes blaTEM, blaSHV, blaCTX-M we used the primers listed in Table 2. PCR included 1 min denaturation (95 °C followed by 30 cycles of 96 °C for 30 s, 62 °C for 30 s, and 72 °C for 30 s and final extension of 72 °C for 10 min. Conditions were identical for other assays except the annealing temperatures which were 55 °C and 44 °C for blaCTX-M and blaTEM, respectively, as described in Refs. 28,29. Amplicons were resolved in 1% agarose gels; for determination of DNA band sizes were used a 100 bp ladder (VWR, Cat No. K180). blaTEM amplicons were purified and sequenced to verify sequence identity (Macrogen Inc, Maryland USA).
Primers used to amplify MexABR genes, pyoverdine receptor genes (fpvA) genes and β-lactamase genes blaTEM, blaSHV, blaCTX-M.
| Gene | Forward primer sequence | Reverse primer sequence | Product size (bp) |
|---|---|---|---|
| mexA | CTCGACCC GATCTACGTC | GTCTTCACCTCGACACCC | 503 |
| mexR | GAACTACCCCGTGAA TCC | CACTGGTCGAGGAGATGC | 411 |
| mexB | TGTCGAAGTTTTTCATTGATAG | AAGGTCACGGTGATGGT | 280 |
| fpvAI | CGAAGGCCAGAACTACGAGA | TGTAGCTGGTGTAGAGGCTCAA | 326 |
| fpvAII | TACCTCGACGGCCTGCACAT | GAAGGTGAATGGCTTGCCGT | 897 |
| fpvAIII | ACTGGGACAAGATCCAAGAGAC | CTGGTAGGACGAAATGCGAG | 506 |
| blaSHV | GCGAAAGCCAGCTGTCGGGC | GATTGGCGGCGCTGTTATCGC | 538 |
| blaCTX-M | GTGCAGTACCAGTAAAGTTATGG | CGCAATATCATTGGTGGTGCC | 538 |
| blaTEM | AAAGATGCTGAAGATCA | TTTGGTATGGCTTCATTC | 425 |
To characterize the prevalence of virulence factors and genes involved in antimicrobial resistance, we collected clinical isolates from various types of wounds. All clinical isolates (n = 51) of P. aeruginosa were pyoverdine producers, as measured by the production of a green fluorescent pigment on CAA agar (Supplementary Fig. 1). Resistance rates (number of resistant isolates) and prevalence (%) of all Pseudomonas aeruginosa isolates against seven antibiotic classes (12 antibiotics) tested are summarized in Fig. 1. In our susceptibility tests, one strain showed resistance to AMI, 0 to GENTA (0%), two to CIPRO (4%), 11 to PIPE (22%), 35 to AMPI (69%), six to CEFTA (12 %), three to CEFE (6%), 27 to CEFO (53%), 33 to TRI (65%), 32 to NITRO(63%), 13 to MERO (25%), 17 to IMI (33%) and, two to CIPRO (12%), three to GENTA (6%), two to AMPI (4%), and 33% to TRI.
Characterization of antibiotic resistance phenotypes in Pseudomonas aeruginosa isolates from Panamá (n = 51). Data shown indicates the number of resistant isolates (red) and prevalence for each isolate (blue) against seven antibiotic classes. Antibiotic key: amikacin (AMI), gentamicin (GENTA), ciprofloxacin (CIPRO), ceftazidime (CEFTA), cefepime (CEFE), cefotaxime (CEFO) piperacillin (PIPE), ampicillin (AMPI), trimethoprim (TRI), nitrofurantoin (NITRO), meropenem (MERO) and imipenem (IMI).
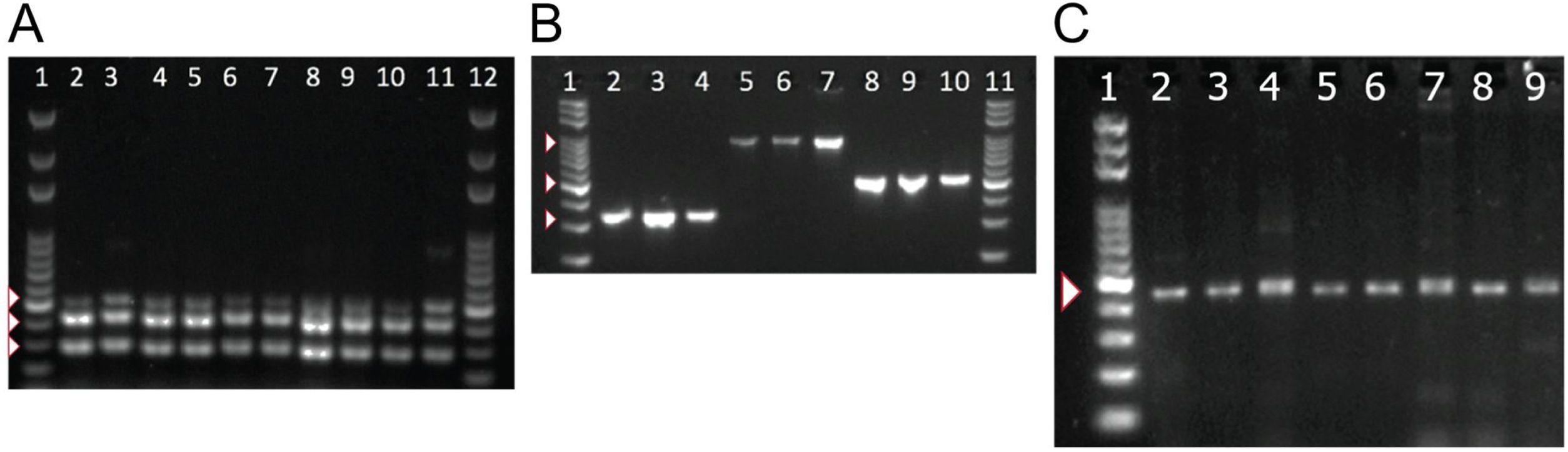
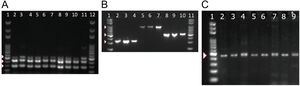
To detect the presence of components of the MexAB-OprM system, we used PCR to amplify gene products for mexA, mexB, mexR. Amplicons for mexA (product size: 503 bp), mexR (product size: 411 bp) and mexB (product size: 280 bp) were detected in all 51 isolates as shown in Fig. 2A.
PCR amplification of MexAB-OprM system, pyoverdine receptors genes, and the β-lactamase gene blaTEM. Detection of mexA, mexB and mexR genes of P. aeruginosa isolates by PCR (A). Lane 1 and 12 corresponds to the molecular weight marker (100 bp DNA ladder). Amplicon sizes: mexA (503 bp), mexB (280 bp) and MexR (411 bp). Positive bands are indicated by white arrowheads. Molecular detection of pyoverdine receptor genes fpvAI, fpvAII and fpvAIII genes from P. aeruginosa isolates by PCR (B). Lane 1 and 11 corresponds to the 100 bp DNA Ladder. Lanes 2-4: fpvA type I (326 bp); Lanes 5-7: fpvA type II (897 bp) and 8-10: fpvA type III amplicons (506 bp). Positive bands are indicated by white arrowheads. PCR Amplification and sequencing of blaTEM-like β-lactamase genes in eight positive isolates of P. aeruginosa (C). PCR detection of the β-lactamase gene blaTEM (425 bp). Lane 1 on panel A corresponds to the 100 bp molecular weight DNA ladder. Positive bands are indicated by a white arrowhead. Multiple sequence alignments showing homology to blaTEM-like genes from E. coli and P. aeruginosa are shown in Supplementary Fig. 2.
Next, we tested for the presence of pyoverdine receptors genes, fpvAI, fpvAII, fpvAIII in all of our isolates and found positive amplification for all of the receptor genes in all 51 strains. Amplicons of 326 bp for fpvAI, 897 bp for fpvAII were detected and a PCR fragment size of 506 bp for the fpvAIII receptor gene as shown in Fig. 2B.
To better understand the virulence of our isolates, we proceeded to test for the presence of blaTEM-like, blaSHV-like, blaCTX-M-like β-lactamase genes that may confer resistance against antibiotics. Among the β-lactamase genes tested, PCR products for a blaTEM-like β-lactamase were detected in only 43% of our isolates (22/51). A PCR product of 425 bp for blaTEM-like gene was detected as shown in Fig. 2C, which was confirmed by DNA sequencing for a select number of isolates (Supplementary Fig. 2). Amplicons for blaCTX-M-like or blaSHV-like β-lactamases were not detected in our isolates.
DISCUSSIONPseudomonas aeruginosa is a Gram-negative bacterium with a capacity to produce a wide range of virulence factors which makes it a MDR pathogen for which there are limited therapeutic options. Therefore, the understanding of resistance mechanisms can facilitate the development of new therapeutics.
In this study, we tested the resistance pattern of P. aeruginosa using antibiotics of the following groups: aminoglycosides (amikacin and gentamicin), fluoroquinolones (ciprofloxacin), other β-lactams (cephalosporins, ampicillins) (ceftazidime, cefepime, cefotaxime, piperacillin, ampicillin), pyrimethamine (trimethoprim), nitrofuran (Nitrofurantoin) and carbapenems (meropenem and imipenem).
In Panama, the Central Reference Public Health Laboratory (LCRSP) of the Gorgas Memorial Institute for Health Studies (ICGES) reviewed the National Antibiotic Resistance Database from 2007 until 2013.30 For the purpose of our study, we chose the Gram-negative bacterium P. aeruginosa because of its medical importance and innate ability to develop antibiotic resistance. In our study, this bacterium was shown to be more susceptible to two aminoglycosides gentamicin (100%) and amikacin (98%), fluoroquinolones (ciprofloxacin, 96%) and two third generation cephalosporins, ceftazidime (88%) and cefepime (94%), as shown in Fig. 1. By contrast, the isolates tested in this study showed increased resistance to antibiotics in the following categories: (i) penicillins (ampicillin 69%, piperacillin 22%); (ii) pyrimethamines (trimethoprim 65%); (iii) nitrofurans (nitrofurantoin 63%), and (iv) third-generation cephalosporin cefotaxime (53%), in contrast to previous reports.30 Resistance values for carbapenems oscillated between 25 and 33% for meropenem and imipenem, respectively.
In the United States, the National Healthcare Safety Network reported that Pseudomonas aeruginosa is the sixth most common nosocomial pathogen and the second most common in ventilator-associated pneumonia in hospitals.31 Nguyen et al.31 reported 9.7% resistance rate to at least one aminoglycoside and 19.3% to at least one carbapenem compared to 2% and 25-33%, respectively, in this study. The reported resistance rate for cephalosporins was 10.3% (cefepime or ceftazidime) compared to 6%, 12% and 53% for cefepime, ceftazidime and cefotaxime, respectively, in this study. Resistance for one fluoroquinolone was reported in 21.6% of isolates compared to 4% in our study. Overall, the MDR index was reported to be 14.2% (previous study), much lower than the 59% observed in the present study. The difference between the two studies could be explained by the presence of intrinsic genetic differences between circulating isolates, which is corroborated by the presence of various resistance mechanisms and/or genes that contribute to virulence and resistance to various drugs representatives of seven antibiotic classes.
The MexAB-OprM efflux system is responsible for the development of resistance to a number of antimicrobials including quinolones, macrolides, tetracyclines, lincomycin, chloramphenicol, novobiocin, and β-lactams.3,6,7,32,33 In our study we detected the presence of genes of the MexABR-OprM efflux system under iron-limiting conditions, as evidenced by the growth of P. aeruginosa under these conditions (CAA medium) and the presence of amplicons for mexA, mexB and mexR by multiplexed standard PCR (Fig. 2A). These results are in line with two previous reports where all components were encoded in the genome and induced by the iron-limiting conditions and their detections indicate that they confer or allow the development of resistance to different antibiotics.12,33–36 The functional expression of each system components and its individual contribution to the resistance phenotype measured is the topic of a follow up study.
Given that iron is key for the development of antibiotic resistance in Pseudomonas aeruginosa, in this bacterium the mechanism of iron uptake is mediated by siderophores, pyocheline and pyoverdine, making them all crucial for the establishment of infection processes.15,37–39 Several studies have shown that increasing concentrations of iron decreased resistance to antibiotics.40,41 Under iron-repleted conditions, P. aeruginosa shows enhanced resistance to the antibiotics tigecycline and tobramycin; the production of pyoverdine was shown to be an important factor for resistance against tigecycline.41,42 In the present study, all 51 clinical isolates were pyoverdine producers (Table 1) and were PCR-positive for all pyoverdine receptor genes, being fpvAI-type the most prevalent type (44%), followed by fpvAIII-type (31%) and fpvAII-type (31%) (Fig. 2B). Our results agree with a previous study where the clinical isolates studied showed a similar prevalence43 meaning that production of pyoverdine increases virulence of the bacteria.38 A previous study associated P. aeruginosa antibiotic resistance with the presence of fpvA genes; the reported prevalence for the receptor genes fpvAI, fpvAII, and fpvAIII were 30%, 20%, and 23%, respectively.44 Recently, a whole genome sequence of two MDR strains revealed as well that gene encoding putative proteins for iron uptake and other virultent factors were detected in both strains. The two siderophores pyochelin and pyoverdine were observed in both MDR strains and their corresponding receptors genes.45 These results agree with those found in this study for all MDR.
Pyoverdine pigment production is ubiquitous amongst β-lactamase-producing strains of P. aeruginosa. Because both pyoverdine and β-lactamase production are necessary for the acquisition of drug resistance capabilities,25,26 we decided to screen our isolates for the presence of ESBLs genes such as blaTEM, blaCTX-M and blaSHV using PCR. Twenty-two strains were PCR positive for the presence of the ESBL blaTEM1-like genes (43%) (Fig. 2C); no PCR products were obtained for blaCTX-M and blaSHV genes. All blaTEM1-positive isolates were highly similar to the blaTEM1 gene of Pseudomonas aeruginosa strain SJP2. An earlier study in P. aeruginosa by Girlich et al.46 reported a 0% prevalence of CTX-M-like and SHV-like genes, and the presence of a gene that coded for a β-lactamaseVEB-1 (blaTEM-like) inserted in an integron; these results are in line with the present study. By contrast, a study by reported a similar prevalence for blaTEM-like genes (26.7%), as well as low prevalences for low-frequency β-lactamases such as blaCTX (17.3%) and blaSHV (10.7%).47 Similarly, a study by Bokaeian et al.48 reported prevalences of 30% for blaTEM-like, 6.6% for blaVHS-like and 0 % for blaCTX-like β-lactamases. Furthermore, certain strains of Pseudomonas and Acinobacter spp. exhibit profound differences in the prevalence of β-lactamase-like genes; in these strains the most prevalent type is blaVHS-like (75%), followed by blaCTX-like (57.5%) and finally blaTEM (15%).23 Together the presence of β-lactamases and the MexAB-OprM efflux pump system greatly contributes to resistance against many classes of β-lactams in P. aeruginosa.7 Our initial screening provided important clues regarding the potential pathogenicity and geographic occurrence of circulating nosocomial strains in Panama.
Different approaches have been used to fight infections caused by P. aeruginosa, from the design of new drugs to metagenomics projects aimed at leading to the discovery of new antibiotics; however, despite such advances, together they have been of limited success41 due to the diversity of resistance mechanisms that allows the bacterium to evade a wide variety of antibiotics. New therapeutic strategies have been developed, i.e. by combining iron-chelators with antibiotics, given the preponderant physiological role of iron in the physiology of this pathogen.42,49–52 The virulence factors identified in our study provides valuable information regarding the prevalence of resistance genes and their potential impact on treatments that exploit the unique physiology of the pathogen. More importantly, this information can be used to better guide future control strategies, understand the evolution and dissemination of strains hoping to prevent the development of resistance in Pseudomonas aeruginosa.