Bacterial pathogens are becoming more and more resistant to antibiotics used commonly by healthcare practitioners. Despite introduction of new antibiotics, empiric treatment of patients with bloodstream infections (BSIs) became a major challenge for practicing physicians.1,2
To depict the susceptibility patterns of BSI pathogens endemic in a referral pediatric hospital, Children's Medical Center (Tehran, Iran), the antimicrobial susceptibility patterns among 3593 patients were reviewed in a 5-year period (2006–2010). Bacterial identification and disk diffusion susceptibility testing were performed, using standard methods.3
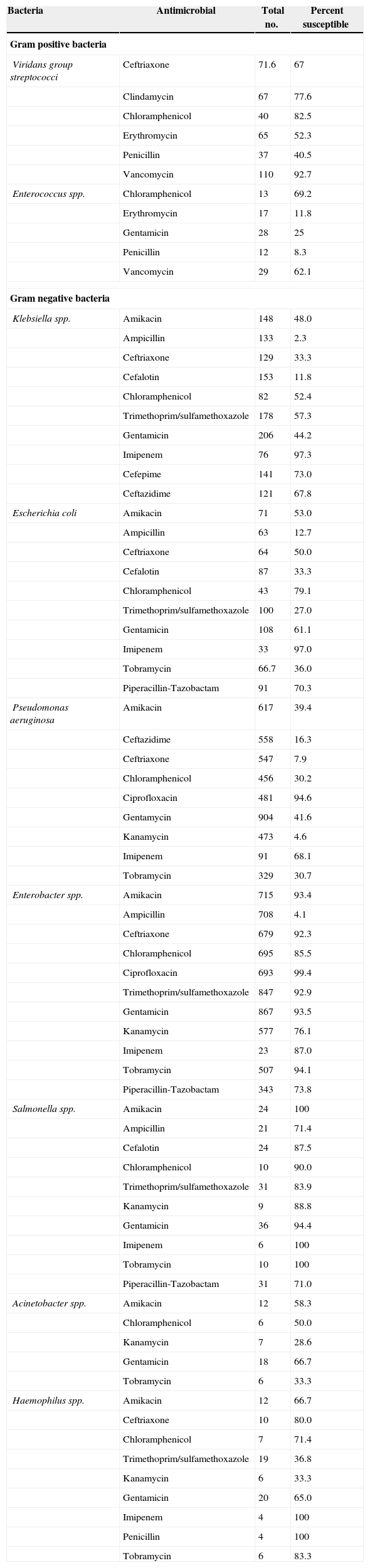
Percentages of Gram positive and Gram negative bacteria were 35% (1265/3593) and 65% (2328/3593), respectively. Enterobacter spp. (25.9%), Pseudomonas aeroginosa (25.5%), and Coagulase-negative staphylococci (CoNS) (23.2%) were the most frequent agents, followed by Staphylococcus aureus (6.2%) and Klebsiella spp. (5.9%), collectively accounting for 87% of all BSI blood isolates cultured. were Staphylococcus spp., followed by Viridans group streptococci, Streptococcus pneumonia, and Entrococcus spp. comprised about 84% of all Gram positive bacteria isolated from blood cultures. Among Gram negative bacteria, Enterbacter spp., Pseudomonas aeruginosa, Klebsiella spp., E. coli, and Salmonella spp. accounted for about 95% of isolates. Table 1 shows the antimicrobial susceptibility pattern of Gram positive and Gram negative bacteria in this study. CoNS are almost always resistant to oxacillin. Furthermore, the rate of susceptibility is very low among S. aureus (13.2%) and S. pneumoniae (7%). Vancomycin showed an acceptable antibiotic effect on CoNS, S. aureus, S. pneumoniae and viridans group Streptococci, with susceptibility rates of 93.2%, 95.3%, 96.4% and 92.7%, respectively. Among staphylococcal isolates S. aureus was more susceptible to clindamycin (82.9% versus 52.9%) and to trimethoprim/sulfamethoxazole (60.9% versus 34.0%) than CoNS. S. pneumoniae was highly susceptible to ceftriaxone (93.9%), cefalotin (96.6%), and vancomycin (96.4%). About half of the Klebsiella spp. isolates tested were resistant to amikacin. The rate of resistance to ampicillin, ceftriaxone and piperacillin-tazobactam was as high as 97.7%, 66.7% and 35.4%, respectively. The rate of E. coli resistance to amikacin was similar to that of Klebsiella spp. On the contrary, Imipenem showed to be quite effective in both organisms (Table 1).
Antimicrobial susceptibility profile of isolated bacteria.
| Bacteria | Antimicrobial | Total no. | Percent susceptible |
|---|---|---|---|
| Gram positive bacteria | |||
| Viridans group streptococci | Ceftriaxone | 71.6 | 67 |
| Clindamycin | 67 | 77.6 | |
| Chloramphenicol | 40 | 82.5 | |
| Erythromycin | 65 | 52.3 | |
| Penicillin | 37 | 40.5 | |
| Vancomycin | 110 | 92.7 | |
| Enterococcus spp. | Chloramphenicol | 13 | 69.2 |
| Erythromycin | 17 | 11.8 | |
| Gentamicin | 28 | 25 | |
| Penicillin | 12 | 8.3 | |
| Vancomycin | 29 | 62.1 | |
| Gram negative bacteria | |||
| Klebsiella spp. | Amikacin | 148 | 48.0 |
| Ampicillin | 133 | 2.3 | |
| Ceftriaxone | 129 | 33.3 | |
| Cefalotin | 153 | 11.8 | |
| Chloramphenicol | 82 | 52.4 | |
| Trimethoprim/sulfamethoxazole | 178 | 57.3 | |
| Gentamicin | 206 | 44.2 | |
| Imipenem | 76 | 97.3 | |
| Cefepime | 141 | 73.0 | |
| Ceftazidime | 121 | 67.8 | |
| Escherichia coli | Amikacin | 71 | 53.0 |
| Ampicillin | 63 | 12.7 | |
| Ceftriaxone | 64 | 50.0 | |
| Cefalotin | 87 | 33.3 | |
| Chloramphenicol | 43 | 79.1 | |
| Trimethoprim/sulfamethoxazole | 100 | 27.0 | |
| Gentamicin | 108 | 61.1 | |
| Imipenem | 33 | 97.0 | |
| Tobramycin | 66.7 | 36.0 | |
| Piperacillin-Tazobactam | 91 | 70.3 | |
| Pseudomonas aeruginosa | Amikacin | 617 | 39.4 |
| Ceftazidime | 558 | 16.3 | |
| Ceftriaxone | 547 | 7.9 | |
| Chloramphenicol | 456 | 30.2 | |
| Ciprofloxacin | 481 | 94.6 | |
| Gentamycin | 904 | 41.6 | |
| Kanamycin | 473 | 4.6 | |
| Imipenem | 91 | 68.1 | |
| Tobramycin | 329 | 30.7 | |
| Enterobacter spp. | Amikacin | 715 | 93.4 |
| Ampicillin | 708 | 4.1 | |
| Ceftriaxone | 679 | 92.3 | |
| Chloramphenicol | 695 | 85.5 | |
| Ciprofloxacin | 693 | 99.4 | |
| Trimethoprim/sulfamethoxazole | 847 | 92.9 | |
| Gentamicin | 867 | 93.5 | |
| Kanamycin | 577 | 76.1 | |
| Imipenem | 23 | 87.0 | |
| Tobramycin | 507 | 94.1 | |
| Piperacillin-Tazobactam | 343 | 73.8 | |
| Salmonella spp. | Amikacin | 24 | 100 |
| Ampicillin | 21 | 71.4 | |
| Cefalotin | 24 | 87.5 | |
| Chloramphenicol | 10 | 90.0 | |
| Trimethoprim/sulfamethoxazole | 31 | 83.9 | |
| Kanamycin | 9 | 88.8 | |
| Gentamicin | 36 | 94.4 | |
| Imipenem | 6 | 100 | |
| Tobramycin | 10 | 100 | |
| Piperacillin-Tazobactam | 31 | 71.0 | |
| Acinetobacter spp. | Amikacin | 12 | 58.3 |
| Chloramphenicol | 6 | 50.0 | |
| Kanamycin | 7 | 28.6 | |
| Gentamicin | 18 | 66.7 | |
| Tobramycin | 6 | 33.3 | |
| Haemophilus spp. | Amikacin | 12 | 66.7 |
| Ceftriaxone | 10 | 80.0 | |
| Chloramphenicol | 7 | 71.4 | |
| Trimethoprim/sulfamethoxazole | 19 | 36.8 | |
| Kanamycin | 6 | 33.3 | |
| Gentamicin | 20 | 65.0 | |
| Imipenem | 4 | 100 | |
| Penicillin | 4 | 100 | |
| Tobramycin | 6 | 83.3 | |
Although bacteriologic culture is the keystone of management of septicemia, culture result takes time; therefore, understanding the regional bacterial susceptibility and pattern of resistance to antimicrobial agents is very important to prepare treatment guidelines. In two previous studies conducted in the same hospital during 1995–2000 and 2001–2005, the prevalence of Gram positive bacteria was reported as 72.0% and 47.6%, respectively.4,5 However, the recent data showed that the Gram negative microorganisms have become much more prevalent. These data also show that the pattern of antimicrobial resistance in Iran is different from other parts of the world. Thus, based on the observed changes therapeutic regimens prescribed by health practitioners in Iran should be modified.
Conflict of interestThe authors declare no conflict of interest.