Infection caused by carbapenem-resistant Klebsiella pneumoniae has become a major healthcare threat and KPC-2 enzyme is a dominant factor mediating carbapenems resistance in K. pneumoniae. This study was designed to determine the genetic environment of blaKPC-2, which prevailed in clinical K. pneumoniae isolates recovered in Huashan Hospital, Shanghai, China. Forty-two clinical isolates were included in this study by blaKPC-2 screening. After multilocus sequence typing and plasmid analyses of PCR-based replicon typing (PBRT), junction PCR, mapping PCR and crossing PCR assays, primer walking, and amplicon sequencing were used to analyze the genetic environment of the blaKPC-2 gene. ST423, ST65, ST977, and ST11 were all detected in KPC-2-producing K. pneumoniae. Two types of blaKPC-2-bearing genetic structure were found: Tn1721-blaKPC-2-Tn3 and Tn1721-blaKPC-2-ΔTn3-IS26; and were carried in IncX and IncFII plasmids, respectively. In conclusion, the genetic environment of the blaKPC-2 gene was diverse and Tn1721-blaKPC-2-ΔTn3-IS26 was dominant in clinical K. pneumoniae isolates in Huashan Hospital. This study sheds some light on the genetic environment and should foster further studies about the mechanism of the blaKPC-2 dissemination.
Carbapenem-resistant Enterobacteriaceae, especially Klebsiella pneumoniae, have emerged as important causes of morbidity and mortality among hospital acquired and long-term care-associated infections such as bacteremia and pulmonary infections.1,2 KPC-2, the most common variant of KPC (K. pneumoniae carbapenemases) enzymes, is a dominant factor mediating carbapenems resistance in Enterobacteriaceae.3
In most countries and regions, such as Europe,4 the United States,5 and Brazil6,7blaKPC-2 is mainly located on Tn4401. It is an active transposable element capable of mobilizing this drug-resistant gene at high frequency among Enterobacteriaceae.8 In Chinese initial reports, the blaKPC-2 gene was located on a Tn3-based transposon, Tn1721, on the plasmid pKP048 (GenBank accession no: FJ628167.2).9 Then, Liu et al. described a truncated form of Tn1721 composed of IS26 inserted into tnpRTn3 which was located on pKPHS2 (GenBank accession no: CP003224.1) from a clinical K. pneumoniae HS11286 (Fig. 1).10 Despite a matching sequence in Tn4401 that exhibits identity to the 3kb blaKPC-2-bearing region within Tn1721, Tn4401 did not carry blaKPC-2 in any of the Chinese isolates up to now.9–11 Obviously, distinct genetic environment of blaKPC-2 suggests that the mechanism to mobilize blaKPC-2 in China is different from that in other regions.
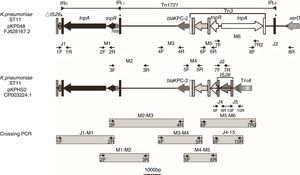
Schematic map of primer annealing sites for junction PCR (J1, J2, J3, J4 and J5); mapping PCR (M1, M2, M3, M4, M5 and M6); and crossing PCR assays. All primers were designed based upon pKP048 (GenBank accession no. FJ628167.2) sequence data, with the exception of the primers corresponding to J4 and J5, which were designed based upon pKPHS2 (GenBank accession no. cp003224.1).
This study reports a nosocomial outbreak of 42 KPC-2-producing K. pneumoniae isolates at our hospital and presented the genetic environment of blaKPC-2 in these isolates using a series of PCR assays and amplicon sequencing instead of the tedious process of plasmid fully sequenced.
Materials and methodsForty-two, non-duplicated, blaKPC-2-postive K. pneumoniae isolates were included in this study by blaKPC-2 screening as described previously,12 after routine identification and antimicrobial susceptibility testing by the Microbiology Laboratory, Huashan Hospital, Fudan University (Shanghai, China) between August 2006 (when the first blaKPC-2-positive clinical K. pneumoniae isolate was detected) and October 2010 (when the blaKPC-2-positive isolates were detected continuously and steadily). The multilocus sequence types (ST) were determined by analyzing seven housekeeping genes (i.e., gapA, infB, mdh, pgi, phoE, rpoB and tonB) as reported previously.13 The results were compared with the MLST databases available at http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html.
Conjugation was performed using donor and recipient Escherichia coli J53 cells mixed at a 1:1 ratio in broth cultures as described previously.13 Transconjugants were selected on MacConkey agar containing imipenem (0.5mg/L) and sodium azide (100mg/L; Sigma Chemical Co.). When resistance plasmid transfer failed in mating experiments, a transformation was used. Plasmids from wild-type isolates were extracted by using a Qiagen Plasmid Midi kit (Qiagen, Germany). E. coli ElectroMAX DH5α competent cells were transformed with the extracted plasmids DNA by electroporation (Micro-Pulser electroporator; Bio-Rad, USA). Transconjugants were selected on MacConkey agar containing imipenem (0.5mg/L). Putative transconjugant colonies were selected and identified by the Vitek system, and further confirmed in a blaKPC-2 PCR assay. Plasmids were classified following the PCR-based replicon typing (PBRT) scheme targeting replicons of the major Inc plasmids.14
Based upon previous reports,9,10 we predicted the genetic environment of blaKPC-2 would be the same as that found in pKP048 and pKPHS2, or present a new structure different from the two structures mentioned above. Junction PCR (J1, J2, J3, J4, and J5) and mapping PCR (M1, M2, M3, M4, M5, and M6) assays were performed for all of the isolates using the relevant primers (Table S1). Junction PCR was mainly used to determine the boundaries of blaKPC-2-bearing genetic structure and to distinguish the sequence of the structure between pKP048 and pKPHS2. Mapping PCR was mainly used for amplifying the backbone of blaKPC-2-bearing genetic structure. Crossing PCR was conducted to cover the whole region bearing blaKPC-2 (Fig. 1). The PCR products were then purified and sequenced. PCR-based primer walking was performed for the isolates that were negative for J1 and J3 using primer 2R and primer 8F, respectively.
The complete pHS062105-3 sequence, and the sequences of the ∼15kb regions surrounding blaKPC-2 in pHS092753 and pHS082416 plasmids were deposited with GenBank under the accession number KF623109.1, KF826293.1, KF724507.1, respectively.
ResultsMLST showed that 34 of these isolates belonged to ST11, five to ST423, two to ST65, and one to ST977. Two Inc groups of plasmids containing blaKPC-2 were detected, namely IncFII and IncX. All blaKPC-2-bearing plasmids found among the ST11 isolates belonged to the IncFII; all the ST423, ST65 and ST977 isolates contained the blaKPC-2-bearing plasmids that belonged to the IncX. Integrating the results of PCR and the sequencing results revealed two types of blaKPC-2-bearing genetic structure (Fig. 2).
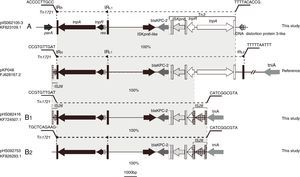
Schematic representations of blaKPC-2-bearing genetic elements classified as either A- or B-type and the sequence bearing blaKPC-2 in pKP048 (GenBank accession no. FJ628167.2). Genes are depicted as arrows according to the direction of transcription. blaKPC-2 is dark gray. Inverted repeats are indicated by color-variable, vertical bars shown in corresponding matching colors: Tn1721 (black), Tn3 (white) and IS26 (gray). Regions sharing identical or near-identical sequence across plasmids are indicated by the gray shading between the representations of different plasmids.
Eight isolates were positive by J2 and M1-M6 assays, but negative by the J1, J3, J4, and J5 assays; this genetic structure Tn1721-blaKPC-2-Tn3 was designated A-type. The sequence from tnpATn1721 to tnpATn3 was the same as that found in pKP048, but without IRL2Tn1721 (Fig. 2). The flanking sequences of all A-type structures identified in this study were indistinguishable. They were located downstream from parA and upstream from a DNA distortion protein 3-like gene. Even the backbone of genetic environment was the same as that found in pKP048, the variant was found for the first time in K. pneumoniae. The plasmids containing blaKPC-2 of A type were found among the ST423, ST65, and ST977 isolates.
B-type (B1- and B2-type), Tn1721-blaKPC-2-ΔTn3-IS26 shared the same blaKPC-2 region as that found on pKPHS2 and the plasmids of this type were all found among the ST11 isolates. Twenty-nine isolates were positive by J1, J4, J5, and M1-M5 assays, but negative by the J2, J3, and M6 assays; the genetic structure (designated B1-type) was represented by pHS082416. The left-most end of Tn1721 was fused with a truncated IS26 element just like the structure of pKPHS2. Five isolates were positive by J4, J5, and M1-M5 assays, but negative by the J1-J3 and M6 assays; this genetic structure designated B2-type revealed no connection between Tn1721 and ΔIS26, and thus differed from B1-type. The upstream sequence of Tn1721 in B2-type was the same as a hypothetical protein found on the K. pneumoniae JM45 plasmid p2 (GenBank accession no: CP006658.1). In addition, the right-most part of intact IS26 connected with tniA in both B1 and B2-types.
DiscussionThe increasing incidence of carbapenem-resistant Enterobacteriaceae has become a great public health concern. One of the key factors contributing to the rapid and wide dissemination of blaKPC-2 is its location on a transposable element.4,8 Compared with the in-depth study of the mechanism of Tn4401 which mobilize the blaKPC-2 gene in Europe and the United States, the relevant research is incomplete in view of the lack of the genetic environment of blaKPC-2 in China. The dissection of the relatively intact region bearing blaKPC-2 is the prerequisite work to further study to elucidate the mechanism.
In Brazil, the large majority of KPC-2-producing K. pneumoniae belong to clonal complex 258/11 (ST258, ST11, ST340, and ST437).6,7,15,16 In our hospital, the first blaKPC-2-positive isolate recovered was ST423; ST65 and ST977 has appeared sporadically. ST11 subsequently emerged and became dominant, just consistent with a previous report from China.17 The IncFII plasmids predominated among the blaKPC-2-positive isolates and was located exclusively in K. pneumoniae ST11. Most isolates could produce transconjugants by mating experiments, suggesting that the spread of blaKPC-2 between different clones of K. pneumoniae might be partly mediated by transmission of blaKPC-2-carrying plasmids in China.
Our analysis just focused on the genetic environment of the blaKPC-2 gene and flanking sequences and presented the two different blaKPC-2-bearing structures in our hospital. Tn1721-blaKPC-2-ΔTn3-IS26 was the dominant one. In methodology, PCR assays and sequencing could be a convenient method to detect the genetic environment of drug-resistant genes instead of plasmids full sequencing as before.18–20
In summary, our study of 42 isolates of K. pneumoniae in Huashan Hospital found the genetic environment of blaKPC-2 to be diverse and Tn1721-blaKPC-2-ΔTn3-IS26 was the dominant genetic structure. The results of this study will assist further studies about the mechanism of horizontal transfer of the blaKPC-2 gene and help controlling the wide dissemination of this cumbersome drug-resistant gene.
Conflicts of interestThe authors declare no conflicts of interest.
We are grateful to Drs. Ming Xu and Stephen H. Gregory (Providence, RI, USA) for their contributions to writing and editing this manuscript. This work was supported by the National Natural Science Foundation of China (Nos. 81071396, 81572031), Shanghai Municipal Commission of Health and Family Planning (No. 20154Y0128).