Community-genotype methicillin-resistant Staphylococcus aureus (CG-MRSA) emerged in the 1990s as a global community pathogen primarily involved in skin and soft tissue infections (SSTIs) and pneumonia. To date, the CG-MRSA SSTI burden in Latin America (LA) has not been assessed.
ObjectiveThe main objective of this study was to report the rate and genotypes of community-genotype methicillin-resistant Staphylococcus aureus (CG-MRSA) causing community-onset skin and soft tissue infections (CO-SSTIs) in LA over the last two decades. In addition, this research determined relevant data related to SSTIs due to CG-MRSA, including risk factors, other invasive diseases, and mortality.
Data sourcesRelevant literature was searched and extracted from five major databases: Embase, PubMed, LILACS, SciELO, and Web of Science.
MethodsA systematic review was performed, and a narrative review was constructed.
ResultsAn analysis of 11 studies identified epidemiological data across LA, with Argentina presenting the highest percentage of SSTIs caused by CG-MRSA (88%). Other countries had rates of CG-MRSA infection ranging from 0 to 51%. Brazil had one of the lowest rates of CG-MRSA SSTI (4.5–25%). In Argentina, being younger than 50 years of age and having purulent lesions were predictive factors for CG-MRSA CO-SSTIs. In addition, the predominant genetic lineages in LA belonged to sequence types 8, 30, and 5 (ST8, ST30, and ST5).
ConclusionThere are significant regional differences in the rates of CG-MRSA causing CO-SSTIs. It is not possible to conclude whether or not CG-MRSA CO-SSTIs resulted in more severe SSTI presentations or in a higher mortality rate.
Staphylococcus aureus is one of the most important human pathogens. It has developed intricate mechanisms to escape the immune system and efficiently invade and damage host tissues, causing multiple clinical syndromes.1 The success of S. aureus as a widely disseminated human pathogen is in part a result of its ability to efficiently colonize the upper respiratory tract and other mucosal and epithelial tissues, which serve as reservoirs for infection. Approximately 20% of the human population can be persistent S. aureus nasal carriers, with an additional 30% subject to intermittent colonization.2 In addition, S. aureus has evolved to be resistant to nearly all classes of antimicrobial agents, promoting the selection and expansion of highly antibiotic-resistant lineages. These can efficiently disseminate into the community and hospital environments and dominate the population structure of S. aureus, causing infections in these settings.3
On a related note, methicillin-resistant S. aureus (MRSA) emerged in the 1960s as an important cause of hospital-associated (HA-MRSA) infections but was confined to health care environments for decades.4 In the 1990s, MRSA found its way into the general population, where it has been primarily involved in skin and soft tissue infections (SSTI) and pneumonia. In some cases, it can be necrotizing.3,4 Nevertheless, HA-MRSA and community-acquired MRSA (CA-MRSA) lineages are genetically distinct.5 The main global HA-MRSA clones are likely clustering, mostly in two clonal complexes (CC5 and CC8), which are also closely related to each other. On the other hand, CA-MRSA clones are more diverse dispersing in many clonal complexes, and only a few are genetically associated with each other.5 HA-MRSA harbors a larger chromosome cassette (SCCmec types I, II, and III), whereas the latter has small mobile genetic elements (primarily SCCmec types IV and V).5 Moreover, most CA-MRSA clones has evolved independently of hospital strains and harbor the genes lukS-PV and lukF-PV, which express a pore-forming cytotoxin Panton–Valentine leukocidin (PVL), absent in HA-MRSA clones.5 Therefore, the major genetic lineages disseminated into the community were originally different from those originating in hospitals. However, the common CA-MRSA have invaded the health care setting, to some extent replacing the canonical HA-MRSA strains.3 In Latin America (LA), MRSA is a common cause of hospital- and community-associated infections ranging from mild to life-threatening diseases.6–11 Despite its role in causing community-acquired diseases, MRSA’s genotypic characteristics isolated from these patients remain poorly understood.
In the early 2000s, the first cases of SSTI and necrotizing pneumonia caused by CA-MRSA strains were reported in LA. An outbreak caused predominantly by an ST30-SCCmec type IV strain was identified among patients in two different hospitals in the metropolitan area of Montevideo, Uruguay and among inmates of two major prisons in the same area.12 Most of these patients presented with abscesses, boils, and cellulitis. Limited information was extracted from the sporadic studies reporting on the molecular epidemiology of CA-MRSA in LA following this outbreak, demonstrating that this genetic lineage had become one of the dominant clones circulating in the community.3 Other community-genotype (CG-MRSA) strains have also been identified as common causes of SSTIs in LA. They seem to be well adapted and widespread throughout the community and are now disseminating within health care settings, often replacing the hospital genotypes.13–17
Although limited information exists on the molecular characterization of CA-MRSA associated with SSTI in LA, more attention has been given to hospital-associated infections. Many studies have demonstrated an important shift in the clonal distribution of HA-MRSA causing invasive diseases, such as bloodstream infections, in the past two decades, with significant differences being detected between countries. Additionally, the major clonal complexes identified in these settings have differed from one another in their antimicrobial resistance, virulence, and mortality rates.18–21 Whether a similar geographical and clonal variability pattern exists for CA-MRSA-causing SSTIs in the LA region is yet to be fully determined. This narrative review aimed to describe data on the available rates of CG-MRSA, defined as MRSA strains harboring SCCmec types IV or V, in community-onset SSTIs in LA. This study also sought to identify additional relevant data on risk factors for CO-SSTIs caused by CG-MRSA, development of secondary invasive disease, and mortality.
MethodsInformation sources and search strategyA systematic review was conducted according to the PRISMA guidelines.22 A search was conducted throughout the Embase, Web of Science, PubMed, Literatura Latino-Americana e do Caribe em Ciências da Saúde (LILACS), and Scientific Electronic Library Online (SciELO) databases for articles published from January 2002 until February 2020, using search terms relevant to S. aureus, SSTI, and names of LA countries. English terms were used when searching all databases, and Spanish and Portuguese terms were used when searching the LILACS and SciELO databases. This study applied the MeSH terms in PubMed, in LILACS as a “subject descriptor,” in SciELO as a “subject,” and as a free text search in Embase. Fig. 2 in Appendix A presents this search strategy. In addition, references cited in eligible studies were visually scanned. Beyond this, gray literature searches were randomly directed to many databases, mainly conference proceedings and theses banks.
Inclusion and exclusion criteriaThis study included all types of observational studies (cross-sectional, case-control, and cohort). The relevant titles and abstracts were added, and their full-text articles were included according to the eligibility criteria developed based on (1) region/country, (2) MRSA definitions (molecular or epidemiological), (3) study design, (4) SSTI definition, and (5) settings. Articles that reported nonhuman isolates, did not provide a clear definition of SSTIs, and those that did not perform MRSA genotyping (at least to the SCCmec level) were excluded. To meet the main objective of this systematic review, SSTIs of community origin, regardless of healthcare risk factors, were considered “community-onset” (CO) infections. Complicated SSTIs were considered invasive diseases, sepsis with organic dysfunction, septic shock, or necrotizing fasciitis. In the case of missing data, the authors were contacted.
OutcomesThe main outcomes of interest were the rate of CG-MRSA in Staphylococcus aureus community-onset SSTIs (CO-SSTIs) and the distribution of major MRSA genotypes. The secondary objective was to discuss relevant data regarding risk factors for CO-SSTIs due to CG-MRSA, development of secondary invasive disease, and mortality.
Data extraction and quality assessmentTwo investigators independently screened the records for eligibility based on titles and abstracts. These investigators also independently assessed each study’s full text, extracted relevant data, and assessed their quality and risk of bias (Table A10). Opinions from the third review author were sought to resolve disagreements. In addition, this study assessed the methodological quality of all included studies using the Joanna Briggs Institute Critical Appraisal Checklist for studies reporting prevalence data.23 This checklist contains nine questions. The quality of the studies was arbitrarily assessed as follows: high quality (>89% of the appropriate items), medium quality (between 67% and 89% of the appropriate items), poor quality (between 44% and 67% of the appropriate items), and very poor quality (<44% suitability). The investigators consensually retained the studies to be included. Disagreements were solved through a discussion with all authors. We used the Preferred Reporting Items for Systematic Reviews and Metanalyses (PRISMA) checklist as a guideline.22,24
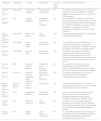
ResultsOut of 429 records identified, 86 studies were selected for full-text assessment (Fig. 2 in Appendix A). Of these, 11 studies were included in the systematic review (Table 1). The included studies were conducted in Argentina (n = 4), Brazil (n = 3), Uruguay (n = 1), Guyana (n = 1), French Guiana (n = 1), and Mexico (n = 1).14,25–34 The number of study participants across the 11 included studies ranged from 13 to 248 (Table 1).14,28
Summary of included studies.
| Reference | Study period | Location | Study design | Exclusive SSTI study? | Aim of study and commentaries |
|---|---|---|---|---|---|
| Pardo et al. (2013)33 | 2003−2006 | Montevideo, Uruguay | Retrospective, observational | No | To describe the relevancy and microbiological aspects of S. aureus isolates from children treated at a tertiary-care; focus on invasive CA-MRSA disease |
| Sola et al. (2008)32 | 2005 | Cordoba, Argentina | Prospective, observational and multicenter | No | To investigate the prevalence of CA-MRSA infections during a surveillance period in 2005. This study was the first report of the prevalence of CA-MRSA in Argentina. Fourteen hospitals participated. The results showed the emergence of CA-MRSA in the community in Argentina |
| De los Monteros et al. (2013)31 | 2006−2008 | Mexico City, Mexico | Cross-sectional | Yes | To determine MRSA prevalence in outpatients presenting with pyodermitis |
| Gelatti et al. (2013)27 | 2007−2008 | Porto Alegre, Brazil | Prospective, observational | Yes | To characterize S. aureus isolates from community-acquired skin and soft tissue infection but searching for CA-MRSA; it did not evaluate the risk factors for CA-MRSA SSTI |
| Bonesso et al. (2014)25 | 2008−2009 | Botucatu, Brazil | Prospective, observational | Yes | To establish the prevalence of MRSA in outpatients attended at a dermatologic treatment center; patients with previous hospitalization and healthcare employees were considered to be at risk to acquired S. aureus SSTI |
| Egea et al. (2014)26 | 2009 | Argentina (several regions) | Prospective, cross-sectional, multicentre | No | To evaluate the molecular and clinical epidemiology of CA- and HA-MRSA in community and healthcare settings. Surgical site infection was not counted as skin disease |
| Furst et al. (2013)14 | 2010−2011 | Santa Fe province and Buenos Aires, Argentina | Prospective, observational, and multicentre | Yes | To establish the prevalence, clinical and molecular attributes of CA-MRSA in adolescents and adults with SSTI |
| Baba-Moussa et al. (2011)30 | Unknown (2010 or 2011) | Cayenne, French Guyana | Case-control | Yes | To establish the association of virulence factors, especially in HIV and non-HIV patients, regarding secondary skin infections |
| Dozois et al. (2015)29 | 2012 | Georgetown, Guyana | Prospective, observational | Yes | The aim was to determine the frequency of MRSA among S. aureus isolates at an emergency department and to identify molecular characteristics of MRSA strains |
| Tomatis et al. (2018)34 | 2013 | Santa Fe, Argentina | Cross-sectional | No | To describe S. aureus strains regarding genotypic study of mecA, SCCmec cassette, PVL and polymorphisms; also, to relate patient data with pheno- and genotypic findings of isolates |
| Souza (2017)28 | 2015 | Niterói, Brazil | Cross-sectional cohort study | Yes | To analyze the prevalence of colonization and cutaneous infection by CA-MRSA in pediatric dermatology outpatients and the factors associated with skin infections |
NR, not reported, NA, not applicable.
Based on three studies performed in Argentina, CO-SSTIs varied between 41 and 90.9% of all identified infections due to CG-MRSA (Table 3).26,32,34
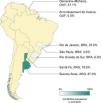
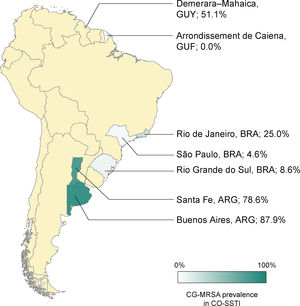
The rates of CG-MRSA in CO-SSTIs are presented in Table 2 and Fig. 1. Most studies included patients of all ages.14,27,29–31,35 The highest percentage of CG-MRSA was 87.9%, which occurred in an Argentinian population aged over 14 years.14 In this study, local regions from which participants were recruited were not mentioned. The predominant MRSA genetic lineages disseminated throughout the community in Argentina were ST30-SCCmec IV and ST5-SCCmec IV.14,26,36 Data extracted from case-control studies conducted in French Guiana showed that the lowest prevalence was 0% (0/55).30 In Guyana, 51% of S. aureus CO-SSTI cases were caused by CG-MRSA, each one characterized as ST8 and SCCmec type IV.29 In Brazil, the highest percentage of CG-MRSA SSTI was 25% in children and adolescents presenting with various baseline dermatological conditions who developed SSTIs.28 For the other two Brazilian studies investigating adult populations in two different regions, Rio Grande do Sul and São Paulo, their respective rates of CG-MRSA were 8.6% and 4.5% (Figure 1).25,27 The small number of cases (n = 5) included in one Brazilian study were mainly associated with ST30-SCCmec-IV strains (n = 3). ST8-SCCmec IV and ST5-SCCmec IV strains caused the remaining two cases.27 In three studies, the characteristics of the isolated MRSA clones were not provided.25,28,31
CG-MRSA percentage in Staphylococcus aureus CO-SSTI.
| Reference | Study period | Inclusion criteria | No. of Staphylococcus aureus CO-SSTI | No. of CG-MRSA SSTI infection (%)a | CG-MRSA clone |
|---|---|---|---|---|---|
| Furst et al. (2013)14 | 2010–2011 | Outpatients ≥14 years, with SSTI and had culture obtained | 248 | 218 (88) | Number of CG-MRSA isolates = 146 |
| ST5-SCCmec IV (24%) | |||||
| Outpatients ≥ 14 years, with SSTI and had culture obtained | ST30-SCCmec IV (48%) | ||||
| SCCmec V (0.7%) | |||||
| (ST not determined) | |||||
| Gelatti et al. (2013)27 | 2007–2008 | Outpatients or patients admitted to the hospital for up to 48 h and presence of superficial cutaneous and soft tissue infections | 58 | 5 (8.6) | Number of CG-MRSA isolates = 5 |
| ST30-SCCmec IV (60%) | |||||
| ST8-SCCmec IV (20%) | |||||
| ST5-SCCmec IV (20%) | |||||
| Bonesso et al. (2014)25 | 2008–2009 | Outpatients of all ages with pyodermitis, diabetes-related infected foot, suppurative osteomyelitis, secondarily infected dermatitis, and secondarily infected traumatic lesions | 66 | 3 (4.5) | The three strains had 81% similarity by PGFE, but there was no association with known CG-MRSA clones. |
| Dozois et al. (2015)29 | 2012 | Outpatients of all ages with SSTI with obtainable purulent material | 47 | 24 (51.1)b | Number of CG-MRSA isolates = 24 |
| ST8-SCCmec IV (100%) | |||||
| Baba-Moussa et al. (2011)30 | Unknown (2010 or 2011) | Outpatients: HIV, non-HIV, and non-infectious dermatosis patients (control) presenting lesions like furuncles or skin infection of a pre-existing non-infectious dermatosis due to S. aureus | 55 | 0 (0) | NA |
| De los Monteros et al. (2013)31 | 2006–2008 | Adults and children outpatients with pyodermitis (impetigo, folliculitis, furunculosis and cellulitis) | 37 | 16 (38) | CG-MRSA clone was not determined, only SCCmec |
| Tomatis et al. (2018)34 (data extracted from outpatients) | 2013 | Outpatients and inpatients (16−65 years) | 13 | 10 (77) | Number of CG-MRSA isolates = 10 |
| Consecutive positive samples | ST30-SCCmec IV (50%) | ||||
| ST5-SCCmec IV (50%) | |||||
| Souza (2017)28 | 2015 | Age ranging from 0 to 19 years old | 16 | 4 (25) | Not determined |
| Non-infectious dermatosis outpatients from public hospital or private clinic |
CO, community onset; SSTIs, skin and soft tissue infections; CG-MRSA, community-genotype methicillin-resistant Staphylococcus aureus; NA, not applicable; CI, confidence interval.
Proportion of CO-SSTI in infections due to CG-MRSA.
| Reference | Study period | Inclusion criteria | No. of CG-MRSA infections | No. of CG-MRSA SSTI (%)a | CG-MRSA clone |
|---|---|---|---|---|---|
| Sola et al. (2008)32 (data extracted from 2005 period) | 2005 | All ages | 22 | 20 (91) | Number of CG-MRSA isolates = 20 |
| Outpatients | ST5-SCCmec IV | ||||
| Single patient with HA-MRSA and CA-MRSA isolates | |||||
| Egea et al. (2014)26 (data extracted from hospitalized patients) | 2009 | All ages | 105 | 43 (41) | Number of CG-MRSA isolates = 43 |
| Outpatients and inpatients | ST5-SCCmec IV | ||||
| Only the first isolate from each patient was evaluated | ST30-SCCmec IV | ||||
| Tomatis et al. (2018)34 (data extracted from outpatients) | 2013 | Adults (16−65 years) | 11 | 10 (91) | Number of CG-MRSA isolates = 10 |
| Outpatients and inpatients | ST30-SCCmec IV (50%) | ||||
| Consecutive positive samples | ST5-SCCmec IV (50%) |
CO, community onset; SSTI, skin and soft tissue infection; CG-MRSA community-genotype methicillin-resistant Staphylococcus aureus.
A Uruguayan cross-sectional study reported that among complicated CG-MRSA invasive infections, none of them were complicated SSTIs (cSSTIs) (0/65).33 The participants were pediatric patients and had invasive infections, as per the authors’ criteria. In an Argentinian study, 27% of hospitalized SSTI patients presented with complicated infections caused by CG-MRSA, mainly comprised of bacteremia and other concomitant infections.26
Risk factors and mortality associated with CG-MRSA CO-SSTIsThe multivariate analysis of an Argentinean study identified two factors independently associated with CG-MRSA in patients with SSTIs (Table A11): the presence of purulent lesions (odds ratio [OR] 3.29, 95% confidence intervals [CI] 1.67, 6.49; p = 0.0006) and age <50 years (OR 2.39, 95% CI 1.22, 4.70; p = 0.01). The presence of necrotic lesions was not statistically significant (OR 0.21, 95% CI 0.22, 2.31; p = 0.57). In the same study, the hospitalization rate was higher when infections occurred due to non-CG-MRSA than CG-MRSA infections (50.5% vs. 33.5%; p = 0.005; CI not provided). However, mortality rates between groups were not significantly different (CG-MRSA, 0.9% vs. non-CG-MRSA, 3.2%; p = 0.16).14
DiscussionThe studies included in our review demonstrated that the effects of CG-MRSA SSTIs in LA greatly differ across the region, with Argentina having the highest rates of infections (88%), followed by Mexico (38%) and Brazil (4.5%–8.6%).14,25,27,31 Published data worldwide also depict significant variation in CG-MRSA SSTIs rates among different regions. A low case proportion has been found in a Chinese multicenter study (2.6%), in which 13 STs were detected.44 In 2017, Australia reported for the first time a predominance (60%) of the CA-MRSA phenotype in CO-SSTIs.45 Meanwhile, published European data reported low MRSA rates in CO-SSTIs, ranging from 0.9% in the Netherlands to 8.3% in France.46 Conversely, in the United States, the percentage of MRSA in CO-SSTI reportedly reached 78% in 2004.47
In contrast to above-mentioned LA countries, Uruguay reported a low infection rate (0.7%) in the outpatient pediatric population, 12 years after identifying the high rates of CA-MRSA phenotype in CO-SSTIs. Clearly, there has been an epidemiological shift in favor of MRSA-causing CO-SSTIs, necessitating additional genotypic analysis.37,38 Interestingly, this change has not occurred in countries that already had significantly higher numbers of CG-MRSA after 2002, such as Argentina and Colombia.14,34,39–43
CG-MRSA has been reported in LA as the causative pathogen of CO-SSTIs since the early 2000s and has been associated with multiple highly successful, widespread pandemic clones, such as those belonging to clonal complexes 30, 8, and 5.12,48,49 In Uruguay and Argentina, the ST30-SCCmec IV lineage carrying the PVL gene, also known as the Southwest Pacific (SW) clone or USA1100, has been particularly dominant.12,15 This lineage was also common in a small series of cases from the southern state of Rio Grande do Sul in Brazil, which borders Uruguay and Argentina.50 Furthermore, the ST30-SCCmec IV lineage seems to be constrained to southernmost LA, whereas in northern countries, such as Colombia and Guyana, the ST8-IV lineage (USA300, LA variant) has been the predominant cause of CA-MRSA infections.16,29,41,51
Studies in Guyana and Colombia revealed that more than 50% of CO-SSTIs were caused by CG-MRSA, mainly USA300-LV and its variant strains without the arginine catabolic mobile element (ACME).29 However, isolated clones in Uruguay and Argentina in the 2000s were different. Although the SW/USA1100 clone was established in Uruguay in the first years after the 2002 outbreak, the same did not occur in Argentina, a border country, in which a variant of the pediatric clone (ST5-SCCmec IVa PVL+) was the major cause of CG-MRSA infections in the early 2000s.15,39 Later, the ST5-SCCmec IV lineage seemed to have been replaced by the ST30-SCCmec IV clone over the years, causing nearly half of the SSTIs infections in Argentina.14 Fernandez et al. observed that the ST30-SCCmec IV lineage had a higher capacity to survive and replicate in the subcutaneous niche than ST5-SCCmec IV. This suggested a potential mechanism associated with this clonal replacement event and major dissemination of ST30-SCCmec IV in the community.18
Countries such as Bolivia, Peru, and Chile have low-to-moderate prevalence of CG-MRSA nasal carriage,52,53 which could partially explain the scarcity of reports on CA-MRSA infections in these countries. In Brazil, the country presenting the lowest CG-MRSA burden in SSTIs, MRSA nasal colonization rates in the general population, even in at-risk populations, have been low.54–56
There remains limited information available on the molecular epidemiology of CO-SSTIs caused by MRSA throughout LA. However, the sporadic reports included in this review have revealed different local events associated with particular genetic lineages that seem to have been selected for different clones in southern versus northern LA. The paucity of data warrants further investigations with a larger number of patients from diverse geographical locations. This would give a deeper understanding of the dissemination of clones highly adapted to the community setting across this vast region. Such information would further investigation into the clinical presentations and outcomes of SSTIs caused by CG-MRSA in LA. These could not be comprehensively analyzed using the data reviewed in our study. Moreover, data regarding risk factors, hospitalization, and mortality rates in CG-MRSA SSTIs were scarce, coming from only a single country (Argentina), where different clones had spread over many years.14,34,48 No broad conclusions could be drawn regarding the severity or mortality of SSTIs caused by CG-MRSA in LA, although they do not seem to significantly differ from statistics of studies conducted in the USA and Australia.14,45,57
ConclusionRegional differences in the rates of CG-MRSA CO-SSTI are substantial. The clonal distribution may vary significantly between southern and northern LA. It is not possible to conclude whether the lineages circulating in LA have resulted in more severe SSTI presentations, higher hospitalization rates, or greater lethality. Therefore, additional clinical studies focusing on the molecular epidemiology of S. aureus isolated from SSTIs in LA are urgently needed.
Author contributionsPaes Leme RC performed the systematic analysis and wrote the manuscript. Bispo PJM wrote the manuscript and reanalyzed the published study data. Salles MJ independently performed the systematic analysis, wrote the manuscript and reanalyzed the published study data.
Conflicts of interestThe authors declare no conflicts of interest.
AcknowledgmentsWe thank Dr. Antônio Carlos Pignatari and the Laboratório Especial de Microbiologia Clínica (LEMC) at Federal University of São Paulo (UNIFESP) for all support.