Multidrug-resistant tuberculosis (MDRTB) is a serious world health problem that limits public actions to control tuberculosis, because the most used anti-tuberculosis first-line drugs fail to stop mycobacterium spread. Consequently, a quick detection through molecular diagnosis is essential to reduce morbidity and medical costs. Despite the availability of several molecular-based commercial-kits to diagnose multidrug-resistant tuberculosis, their diagnostic value might diverge worldwide since Mycobacterium tuberculosis genetic variability differs according to geographic location.
Here, we studied the predictive value of four common mycobacterial mutations in strains isolated from endemic areas of Brazil. Mutations were found at the frequency of 41.9% for katG, 25.6% for inhA, and 69.8% for rpoB genes in multidrug-resistant strains. Multimarker analysis revealed that combination of only two mutations (“katG/S315T+rpoB/S531L”) was a better surrogate of multidrug-resistant tuberculosis than single-marker analysis (86% sensitivity vs. 62.8%). Prediction of multidrug-resistant tuberculosis was not improved by adding a third or fourth mutation in the model. Therefore, rather than using diagnostic kits detecting several mutations, we propose a simple dual-marker panel to detect multidrug-resistant tuberculosis, with 86% sensitivity and 100% specificity. In conclusion, this approach (previous genetic study+analysis of only prevalent markers) would considerably decrease the processing costs while retaining diagnostic accuracy.
Tuberculosis (TB) remains an important public health concern as about one third of the world population is asymptomatically infected with latent Mycobacterium tuberculosis.1 Multiple health campaigns are being addressed to diminish the incidence of the disease, but TB elimination remains a challenge, due to the emergence of clinical forms of multidrug-resistant strains of M. tuberculosis,1 that are resistant to at least isoniazid (INH) and rifampicin (RIF), the first-line antitubercular (anti-TB) drugs. Beyond increasing TB incidence, mortality, and the rate of initial treatment failure, multidrug-resistant tuberculosis (MDRTB) also increase the cost of TB treatment.1 For example, while a 6-month regimen of INH+RIF (plus pyrazinamide and streptomycin or ethambutol in the first two months) will cure most cases of non-resistant TB, MDRTB-patients will need at least 12–24 months of therapy with more toxic second-line drugs.1,2 In addition, MDRTB accounts for up to 84% of the “retreatment” cases of TB.1 With this perspective, early diagnosis appears to be the best strategy to control MDRTB.1
Conventional culture-based drug-susceptibility testing (DST) for TB can take as much as 90 days to complete, due to the slow growth of M. tuberculosis.3 Since diagnostic delay is an arduous obstacle to effective MDRTB care, development and implementation of molecular approaches for rapid detection of MDRTB are needed for attenuating the MDRTB burden. Towards that end, many studies have investigated gene mutations that confer drug resistance in M. tuberculosis.4–6 As a result, many commercial tests have been developed to detect MDRTB and, consequently, the WHO recommends to all governments the gradual implementation of evidence based commercial tests using molecular techniques for diagnosis and control of MDRTB.1
Overall, these tests aim to detect mutations in the mycobacterial genes involved in drug metabolism, which may confer phenotypic changes associated with antibiotic resistance. In this way, INH resistance is frequently associated with mutations in the catalase-peroxidase enzyme (encoded by the katG gene and involved in metabolic activation of the drug),5 and/or in the enoyl-ACP reductase gene promoter (inhA), required for mycobacterial cell wall biosynthesis.7 Likewise, resistance to RIF is strongly related to mutations in a region of 81bp in the rpoB gene (encoding the beta subunit of the RNA polymerase, the drug target),3,8 resulting in decreased affinity of RIF for the active centre of the enzyme.9 Mutations in several other genes may also lead to INH or RIF resistance but are less frequent.10–12 Thus, common gene markers for MDRTB are S315T in katG, −15C/T (CGT-CAT) in the operator region of inhA, and H526D and S531L in rpoB.4,5
Diagnostic parameters, such as sensitivity and specificity, of commercially available diagnostic tests are poorly understood in a worldwide context.1 This is of interest since there is evidence that geographic location influences on the strain-to-strain M. tuberculosis genetic variability.13–15 In this context, current gold-standard molecular biomarkers to diagnose MDRTB might not be found at reasonable frequencies to guide anti-TB drug choice in neglected regions such as Brazil. Moreover, it is even possible that non-trivial mutations could turn out to be novel biomarkers for MDRTB diagnosis in such regions. In this regard, we genotyped four mycobacterial SNPs (single nucleotide polymorphisms) in isolates from TB patients of an endemic region of Brazil to investigate their predictive value to diagnose MDRTB, and using these new reference standards as gold-standard. Our aim was to test and validate a rapid and cost-effective real-time PCR assay for detecting RIF and INH resistance.
MethodsIsolation, identification and drug sensitivity tests of M. tuberculosisM. tuberculosis isolates were considered as MDRTB when resistant to at least INH and RIF (worldwide used as first-line antitubercular drugs), and as sensitive when susceptible to all anti-tubercular drugs. Therefore, mono-resistant strains for INH or RIF, as well as other poly-drug-resistance not including both INH and RIF were not considered for further experiments in our study. These isolates were randomly selected and collected from a repository located at the Central Public Health Laboratory “Prof. Gonçalo Muniz” (LACEN-BA), state of Bahia, Brazil. All isolates from the collection come from sputum of local patients which completed a demographic and clinical questionnaire after signing an informed consent form. All procedures were approved by the Human Ethical Committee of the Universidade Estadual de Santa Cruz (UESC; Ilhéus, Brazil), under protocol number 098/07.
The selected samples were cultivated on Löwenstein-Jensen agar (LJ). Biochemical tests, including nitrate reduction, niacin test and 68°C catalase inhibition were conducted in order to classify these strains according to the Manual of Tuberculosis Bacteriology.16 Drug sensitivity tests to INH, RIF, ethambutol (EMB), pyrazinamide (PZA), and streptomycin (SM) were performed using the Canetti (1969) multiple proportional dilution method.17 The standard strains M. tuberculosis H37Rv and 636 were used as sensitive and MDRTB controls, respectively, as recommended by the Brazilian Ministry of Health.16 The minimum inhibitory concentrations (MIC), defined as the lowest drug concentration showing complete inhibition of bacterial growth, were as follow: INH≤0.2μg/mL, RIF≤1μg/mL, EMB≤5μg/mL, SM≤2μg/mL, and PZA≤25μg/mL. The criterion for drug resistance was growth of ≥1% of the bacterial population on media containing the critical concentration of each drug.
Genotyping assay and amplificationMycobacterial genomic DNA was extracted from the cultured strains according to Van Soolingen et al.18 The quantity and purity was determined measuring spectrophotometric signals at 260nm and 280nm.
Assays were performed, in accordance to Espasa et al.,19 in a spectrofluorometric thermal cycler (ABI Prism 7500, Applied Biosystems®, Carlsbad, CA, USA), using the primers and probes summarized in Table 1. The Mycobacterium tuberculosis Complex-Specific Insertion Sequence IS6110 was also amplified as internal control. Five amplification reactions were performed for each studied sample. Briefly, the reaction contained: 1X Master Mix solution, 500nM of each primer, 200nM of fluorogenic 5′ exonuclease probes (Taqman®, Applied Biosystems®, Foster City, CA, USA), and 4μL (20ng/μL) of sample in 25μL of reaction volume. The amplification settings were carried out according to the manufacturer's instructions.
Primers and probes designed to detect mutations included in the study.
| Gene (codon) | Primers | Probes |
|---|---|---|
| IS6110a | 5′-GGATAACGTCTTTCAGGTCGAGTAC-3′ 5′-TCGCCTACGTGGCCTTTG-3′ | 5′-GGATAACGTCTTTCAGGTCGAGTAC-3′ |
| katG (315) | 5′-GGGCTGGAAGAGCTCGTATG-3′ 5′-GGAAACTGTTGTCCCATTTCG-3′ | 5′-CGACCTCGATGCCGCTGGTGAT-3′ 5′-CGACCTCGATGCCGGTGGTGAT-3′ |
| inhA (−15) | 5′-CACGTTACGCTCGTGGACAT-3′ 5′-CAGGACTGAACGGGATACGAA-3′ | 5′-AACCTATCGTCTCGCCGCGGC-3′ 5′-AACCTATCATCTCGCCGCGGC-3′ |
| rpoB (526) | 5′-ACCGCAGACGTTGATCAACAT-3′ 5′-GGCACGCTCACGTGACAG-3′ | 5′-CGCTTGTGGGTCAACCCCGA-3′ 5′-CGGCGCTTGTAGGTCAACCCC-3′ |
| rpoB (531) | 5′-ACCGCAGACGTTGATCAACAT-3′ 5′-GGCACGCTCACGTGACAG-3′ | 5′-AGCGCCGACAGTCGGCG-3′ 5′-CAGCGCCAACAGTCGGCG-3′ |
Analysis of the results was performed using the Sequence Detection System software (Applied Biosystems®) and was based on two parameters: the cycle threshold (CT), for which the detection of fluorescence reaches the threshold, and the cumulative fluorescence signal (CFS) of each probe at the end of 40 amplification cycles.
Statistical analysisPearson chi-square test was applied to evaluate differences in genotypic frequencies between sensitive and resistant isolates. Discriminant analyses were performed to build predictive models of INH and RIF resistance based on genotyping data of the four genetic markers used, alone or in combination. Comparison of the observed genetic frequencies was performed using UNPHASED software (v.3.1.7).20 Statistical measures of sensitivity and specificity were performed using IBM SPSS Statistics version 20 (SPSS Inc., Armonk, NY, USA) with a 95% confidence interval. The p-value significance was always set at 5% (p<0.05).
ResultsFirst, a confirmatory sensitivity test was performed on a total of 51 resistant and 50 sensitive catalogued strains (Table 2). From those, 43 (84.3% of resistant-strains) were considered as MDRTB strains exhibiting resistance to both INH and RIF, and were selected for further experiments. Six of the strains showed drug-resistance to only one antibiotic (INH), and only two exhibited multi-resistance other than INH and RIF together. The other 50 strains, catalogued as sensitive, were in deed confirmed to be sensitive to all five antibiotics tested, and were used as negative controls.
As expected,4,5 none of the studied mutations was found in drug-susceptible strains (Table 3). Although this finding suggests that absence of the analyzed katG, inhA, and rpoB mutations is indicative of non-MDRTB strains with 100% specificity, it is noteworthy that a small number of MDRTB samples (14%, n=6) also lacked these mutations (Table 3). Thus, our data reveal that the absence of these common katG, inhA, and rpoB mutations does not necessarily assure non-MDRTB.
Frequency of mutations in katG, inhA and rpoB genes of Mycobacterium tuberculosis strains.
| Mutation (gene) | Frequency in susceptible strains (n=50)a | Frequency in MDRTB strains (n=43)a | X2b | p-Valuec |
|---|---|---|---|---|
| S315T (katG) | 0 (0) | 41.9% (18) | 25.953 | <0.001 |
| −15C/T (inhA) | 0 (0) | 25.6% (11) | 14.507 | <0.001 |
| H526D (rpoB) | 0 (0) | 7.0% (3) | 3.605 | 0.095 |
| S531L (rpoB) | 0 (0) | 62.8% (27) | 44.239 | <0.001 |
| None of above | 100% (50) | 14.0% (6) | – | – |
Conversely, we found that the occurrence of any of the mutations translates identification of MDRTB. Among the 43 strains studied for MDRTB, 18 displayed the S315T (katG) mutation, 11 had the −15C/T (inhA) whereas, 3 and 27 strains showed H526D and S531L polymorphisms, respectively, in rpoB (Table 3). Although displaying 100% specificity, sensitivity to detect MDRTB varies according to the identified mutation. Mutations with greater sensitivity (62.8%) were S531L (rpoB), followed by S315T (katG) and −15C/T (inhA), with 41.9% and 21.6%, respectively (Table 3). The H526 mutation (rpoB) showed only 7% sensitivity without statistically significance to detect drug resistance.
Given that bacteria may reduce effectiveness of a drug through various biological mechanisms at the same time, we addressed whether diagnosis sensitivity could be greater than 62.8% (displayed by the single marker S315T katG) in prediction models including multiple mutations. In this regard, we structured four models of two or three SNPs, as summarized in Table 4 (note that combinations with H526 from rpoB were excluded due to the lack of statistical significance to detect drug resistance). Among them, only model 1 (S315T katG+−15C/T inhA) model did not reach the “basal” sensitivity of 62.8%. The gain in sensitivity of model 2 (S531L rpoB+−15C/T inhA) was only about 2.3%, when compared to the best single marker. In contrast, models 3 (S315T katG+S531L rpoB) and 4 (S315T katG+S531L rpoB+−15C/T inhA) showed a considerable gain in diagnosis sensitivity, by increasing this parameter by 23.2%. Using the last two models, we identified 37 out 43 MDRTB strains carrying at least one of the analyzed markers. Thus, taken in consideration that both models have identical diagnostic predictive values, this suggests that it is unnecessary to genotype three of the studied polymorphisms to achieve greater sensitivity.
Predictive profile of polymorphisms in katG, inhA and rpoB genes related to multidrug resistance in Mycobacterium tuberculosis strains.
| Genes | Polymorphisms | Statistical models | Frequency in resistant strainsa (n=43) |
|---|---|---|---|
| katG or inhA | S315T & −15C/T | Model 1 | 60.5% (26) |
| inhA or rpoB | −15C/T & S531L | Model 2 | 65.1% (28) |
| katG or rpoB | S315T & S531L | Model 3 | 86.0% (37) |
| katG or inhA or rpoB | S315T & −15C/T & S531L | Model 4 | 86.0% (37) |
It is evident that strains, as well as type of mutations and frequency of them in mycobacterial genome, vary according to the geographical region.21,22 Consequently, since the accuracy of current molecular diagnostic tests to identify MDRTB are based on worldwide polymorphisms frequencies, performance indicators for these diagnostic approaches in a country-by-country basis may be controversial. For example, the S315T (katG) polymorphism shows a wide variation in their incidence: although it was detected in a 100% of resistant strains in countries such as Turkey, Canada, and France,23–25 in our study we found a lower frequency of 41.9%, more similar to those obtained in another study in Brazil (60%) and in places such as Russia (77%), Syria (40%) or Taiwan (51%).13,26–28 Likewise, the −15C/T (inhA) polymorphism, described as the one most frequent globally for the inhA gene promoter,5,29 has an observed frequency varying between 15% in China30 to 32% in Syria,13 and 25.6% in this study. Also, the frequency of 62.8% for S531L (rpoB) found in this work was consistent with other studies, considering that published rates range between 40.1% and 82.4%.31–34 In the same way, H526D (rpoB) polymorphism, with a found frequency of 7% in this work amply oscillates worldwide, with values ranging from 5.9% to 40%.31,34 Therefore, great frequency variations are evident between countries, making it necessary to study locally the prevalence of the canonical mutations for each geographical region in order to figure out which polymorphisms have the best predictive accuracy.
As early diagnosis is the best strategy to control MDRTB, several molecular methods to detect drug resistance in M. tuberculosis are currently widely employed.35 For that, a number of commercial kits to identify MDRTB are available, such as Xpert MTB/RIF® (Cepheid, Sunnyvale, CA, USA) and INNO-LiPA Rif.TB (Innogenetics N.V., Gent, Belgium), among others, which detect resistance to only rifampicin not to isoniazid.36,37 The rational for assessing only mutations associated RIF-resistance is that some reports have suggested that RIF-resistance seems to serve as a surrogate marker for MDRTB detection,38,39 as almost all RIF-resistant strains were also INH-resistant. In our study, we found also that 100% of RIF-resistant strains were also INH-resistant (Table 2). Nevertheless, this finding depends of the endemic strains of M. tuberculosis in each geographical region,21,22 as discussed above. In this way, some reports from the Punjab region of India40,41 have shown a high incidence of RIF-monoresistant strains (22%) that would be falsely diagnosed as MDRTB using these type of test. The Xpert® MTB/RIF assay, much cheaper than INNO-LiPA Rif.TB (approximately US$17 and US$45 per test, respectively),42,43 has been recommended by the World Health Organization for the detection of MDRTB from 2010, and is exhaustively implemented worldwide.1,44 However, only a limited number of tests can be performed per day, making it necessary to acquire several machines per laboratory. Furthermore, the cost per test,45 even if negotiated for public institutions by the FIND organization43 (Foundation for Innovative New Diagnostics, Geneva, Switzerland) (Table 5), makes our system an interesting cost/benefit diagnostic tool for our local area. In addition, these kits have sensitivity and specificity to detect drug resistance similar to those shown in this study36,37,45 (Table 5).
Comparative parameters of molecular diagnosis of Multidrug Resistant Tuberculosis between this study and published data for Xpert MTB/RIF.
| Analytical variables | This work | Xpert MTB/RIFa |
|---|---|---|
| Sensitivity | 86.0% | 89.2–91.4% |
| Specificity | 100.0% | 98.0–98.7% |
| No. of polymorphisms studied | 2 | 5 |
| Chemotherapy resistance studiedb | INH, RIF | RIF |
| Time (DNA extraction+PCR reaction) | 3h | 2h |
| Test/day (considering a work day of 8h) | 20–30 | 8–16c |
| Price of 1 test | US$ 8.53 | US$ 9.98–16.86d |
Source: Chang K, Lu W, Wang J, et al. Rapid and effective diagnosis of tuberculosis and rifampicin resistance with Xpert mtb/rif assay: A meta-analysis. J Infect 2012; 64(6): 580–588.45
http://www.finddiagnostics.org/about/what_we_do/successes/find-negotiated-prices/xpert_mtb_rif.html.43
Recently, there has been reported a more accurate commercial kit, the GenoType MTBDRplus (Hain Lifescience, Nehren, Germany) which has higher sensitivity than competitors (up to 94.4%) and could detect even resistance to both INH and RIF.46 Nevertheless, it requires the analysis of up to 10 markers to diagnose MDRTB, whereas Xpert® MTB/RIF and INNO-LiPA Rif.TB use 5 and 4, respectively,42,43 increasing even more the processing costs (between €42 and €86 per test).47 In our study, the proposed analysis of polymorphisms to diagnose MDRTB in the Bahia state of Brasil, has a high sensitivity (86%) even if only two genetic markers are analyzed.
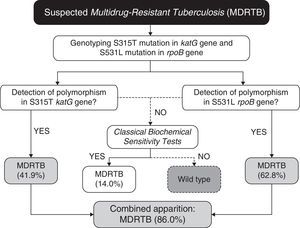
In our case (Bahia state of Brazil), after identification of the prevalent polymorphisms, the protocol for MDRTB analysis, (Fig. 1) would consist in extracting the DNA from M. tuberculosis directly from sputum samples. Then the samples would be subjected to genotyping for the detection of polymorphisms at S531L rpoB and S315T katG genes. Presence of S531L (62.8% of incidence in endemic mycobacteria) or S315T (41.9%) mutations in the samples would be diagnosed as MDRTB (86% of cases, carrying anyone of the two mutations). The specificity of 100% coupled with high accuracy, easy operation, and low cost (due to the smaller number of markers used), makes it a great method to be used in routine diagnosis of MDRTB. Our work shows that combining a few markers, without the need of an extensive panel of analysis, is sufficient to improve the accuracy for the detection of both RIF- and INH-resistance in our region.
Our results came as no surprise, considering that most RIF-resistance mutations occur close to the S531 residue,48 and almost all RIF-resistant strains are also INH-resistant.38,39 Moreover, the S315T mutation is the most frequently found in INH-resistant strains that are also MDRTB, occurring in approximately 40% of all cases.49
For the small percentage of false-negative samples (14%), conventional culture and drug sensitivity techniques can be performed as confirmatory tests, as 86% of the multidrug-resistant population will be previously diagnosed in both effective and early-time manners. In this context, even if further studies should be conducted to establish other markers to increase even more the sensitivity of the proposed algorithm (Fig. 1), taken together our results have shown that in order to provide a fast, sensitive and low-cost diagnosis, the proposed PCR-based protocol would be sufficiently appropriate for our geographical region. As demonstrated previously by Madania et al.,13 in Syria, a small set of probes detecting prevalent mutations can be used in real time PCR in order to detect most strains carrying RIF- and INH-resistance. In the same way, we propose that our algorithm can be revised and implemented in other countries besides Brazil, by conducting a simple genetic background evaluation of their endemic populations of M. tuberculosis.
Conflicts of interestThe authors declare no conflicts of interest.
We thank the Laboratório Central da Bahia (LACEN-BA) for providing isolates and drug susceptibility testing for M. tuberculosis. We also thank Dr. Daniella Oliveira Campo for the help during this research. This study was funded by the Fundação de Amparo a Pesquisa do Estado da Bahia (FAPESB) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). L.A.V.M and A.M. have post-doctoral fellowships from the Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).