Epidemiological studies are important tools to assess the diversity of Brucella isolates and to estimate their epidemiological relationship among isolates from different geographical origins. In this study the MLVA16 (multiple-locus variable number tandem repeat analysis based on 16 loci) was employed to investigate the diversity of Brucella spp. Isolated from humans and animals for epidemiological purposes and to determine the most common Brucella genotypes in Iran.
MethodsWe designed a molecular-based study to evaluate the potential reservoirs of human brucellosis. After isolation and identification of 54 Brucella spp human and animal specimens from three regions of Iran, bacterial genomic DNA was extracted MLVA with three panel was used for the genotyping of isolates. The size of PCR products were analyzed and converted to repeat unit numbers using a published allele numbering system and data set was imported into Bionumerics.
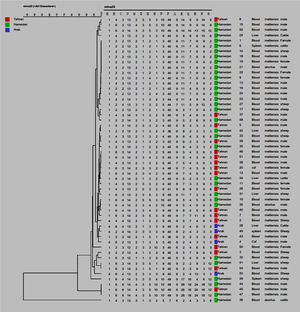
ResultsThree isolates (5.55%) were identified as Brucella abortus and 51 (94.44%) as Brucella melitensis. Two isolates of Brucella abortus were from humans and one from an animal. Thirty-four Brucella melitensis isolates were from humans and 17 from animals. Using MLVA16-genotyping, 54 isolates with genetic similarity coefficient of 80% were divided into 46 genotypes and 22 genotypes were represented by a single isolate, while 4, 2, 1 and 2 genotypes were represented by 2, 3, 4 and 7 isolates, respectively. The most prevalent genotype was represented by 14 isolates. There were two other frequent genotypes each represented by seven isolates, among which only one was restricted to a geographic region. Discriminatory power for each locus was determined in this study and panel 2B shows the high discretionary power [Bruce04 (0.837), Bruce30 (0.806), Bruce 09 (0.787), Bruce 07 (0.772), Bruce16 (0.766)].
ConclusionMLVA16 analysis of 54 Brucella isolates showed high level polymorphism in their genotypes. Only two genotypes, each observed in seven isolates, were related to one another and only one of these genotypes were found in to two separate regions.
Brucellosis is a zoonotic bacterial disease caused by the genus Brucella, which is transmitted through direct or indirect contact to animals.1 Based on differences in pathogenicity, phenotypic characteristics and hosts, the genus Brucella is classified into 11 species.2,3 Four species, including Brucella abortus, Brucella melitensis, Brucella canis and Brucella suis are known to infect humans. Enormous economic losses and public health problems results from abortion, infertility in livestock, weak offspring, decreased milk production, and morbidity in humans. Although brucellosis has been eradicated in the USA, Canada, North Europe, and Australia, this infection is still highly prevalent in central Asia, Middle East, Mediterranean region, Africa, and Latin America.4 Iran is an endemic area for brucellosis and there is a major risk of Brucella transmission from eastern and western neighboring countries such as Iraq, Pakistan, and Afghanistan due to lack of high quality veterinary services for controlling animal diseases.5,6 Although, 500,000 human cases of brucellosis are annually reported worldwide, some cases remain undetected or neglected.7 Key approaches for preventing brucellosis in humans include controlling the disease in animals using epidemiological studies to assess diversity among strains, and estimating the epidemiological relationship between isolates from different geographical origins.8,9
Classical phenotyping methods, such as serotyping, phage typing, metabolic profiling and sensitivity to dyes have been employed for Brucella subtyping; however, these techniques are only available in reference laboratories, have a limited discriminatory power, require manipulation, and lack any standardized interpretation of this method cause difficulties.10
Based on these limitations, bacterial typing has shifted towards molecular identification by which epidemiological relationships are investigated among isolates. Thus far, several molecular typing methods, such as random amplified polymorphic DNA (RAPD)-PCR, amplified fragment-length polymorphism (AFLP), pulsed field gel electrophoresis (PFGE), polymerase chain reaction restriction fragment length polymorphism (RFLP), multiple-locus sequence typing (MLST) and multiple-locus VNTR (variable number tandem-repeats) analysis (MLVA) have been introduced. MLVA method is considered to be an effective and rapid tool for monitoring variations of copy numbers of tandem repeat units (TRs) with a high discriminatory power.11 This method is not only used in outbreaks and epidemiological and trace-back investigations, but also in confirmatory laboratories or screening of food-borne acquired infections. TR sequences are multiple alleles that can be presented on a single locus and be easily determined through agarose electrophoresis or capillary electrophoresis based on their size differences.12 MLVA has been proven to be a good technique for the assessment of pathogenic bacteria, such as Brucella, that display very little genomic diversity. MLVA schemes with 21, 15 and 16 loci (MLVA-21, MLVA-15 and MLVA-16, respectively) have been presented for discriminating Brucella spp. MLVA-16 has been proposed by Al-Dahank et al. with eight minisatellite markers (panel1: Bruce-06, Bruce-08, Bruce-11, Bruce-12, Bruce-42, Bruce-43, Bruce-45, and Bruce-55) for identification of bacteria spp and eight microsatellite markers (panel 2A: Bruce-18, Bruce-19, Bruce-21 and panel 2B: Bruce-04, Bruce-07, Bruce-09, Bruce-16 and Bruce-30) for screening the most common genotypes and classification of isolates to subspecies.13 The genetic diversity of Brucella isolated from human and animal specimens has not yet been investigated in Iran. The main objectives of this study was to employ the MLVA-16 assay to investigate and assess the diversity of Brucella isolates for epidemiological purposes and to determine the most common genotypes of Brucella isolates in Iran.
MethodsEthical statementEthical Committee of Iran University of medical science approved this study under the ethics code R.IUMS.REC 1396.33003.
Study design and sample sizeThis study is a molecular-based and cross-sectional study to evaluate the possible reservoirs of human brucellosis in Iran.
Isolation and determination of Brucella sppThe clinical samples were collected from humans and animals suffering from brucellosis. Culture isolation of Brucella spp was achieved by agar supplemented with 10% horse serum. The inoculated plates were incubated at 37 °C aerobically for 3–5 days. Single colonies were analyzed microscopically using Gram staining and biochemical (urease, oxidase and catalase) and motility tests were carried out for bacterial identification. Colonies confirmed as Brucella spp were subjected to PCR methods for species-level identification.
Preparation of Brucella DNA and amplification of lociDNA of Brucella was extracted using the PCR template preparation Kit (Roche Diagnostics, Germany). MLVA genotyping was performed as previously described by Flech et al. and completed by Al Dahouk et al. This method includes three panels named panel1, panel 2A and panel 2B. The 16 primer pairs were divided into three groups. Panel1 consists of eight minisatellite loci (Bruce 06, 08, 11, 12, 42, 43, 45, 55), while panel 2A (Bruce18, 19, 21) and panel 2B (Bruce 04, 07, 09, 16 and 30) consisted of three and five minisatellite loci, respectively. PCR amplification was performed in a total volume of 15 µl containing 1 ng of DNA, 1X PCR reaction buffer, 1U of Taq DNA polymerase, 200 mM of each deoxynucleotide triphosphate, and 0.3 mM of each flanking primer. Amplification was performed in an Eppendrof termocycler as follow: an initial denaturation step at 96 °C for 5 min, followed by 30 cycles of denaturation at 96 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 70 °C for 1 min. The final extension step was performed at 70 °C for 5 min. Five microliters of amplification product was loaded on 2.5% agarose gel and run under a voltage of 8 V/cm. Gel images were then recorded.
Analysis of MLVA dataThe sizes of PCR products were analyzed and converted to repeat unit numbers using a published allele numbering system and the data set was imported to Bionumerics. For quantification of polymorphism at each locus, Hunted and Gaston diversity index (HGDI), available on the human protein HPA website (http://www.hpa-bioinformatics.org.uk/cgi-bin/DICI/DICI.pl) was used. Categorical coefficient and unweighted pair group methods were applied for clustering analysis. Minimum spanning trees were also constructed.
Statistical analysisStatistical analyses were conducted using the SPSS version 18, (Inc, Chicago, IL, USA).
ResultsCharacteristics of patients and Brucella isolatesA total of 54 Brucella spp. were respectively isolated from blood (62.96%) and cerebrospinal fluid (3.70%) of humans, as well as blood (16.66%), spleen (5.55%) and liver (11.11%) of animals. Human patients included 26 men (48.14%) and 10 women (18.51%) and animals included 15 sheep (27.77%) and three cattle (5.55%). Isolates were collected from three states of Iran, including Hamedan (59.25%), Arak (7.40%), and Tehran (33.33%). All isolates were identified either as Brucella melitensis or Brucella abortus. Three isolates (5.55%) were identified as Brucella abortus and 51 isolates (94.44%) were identified Brucella melitensis. The mean age of the 36 patients was 45, ranging from 6 to 80 years-old and male to female ratio was 2.6.
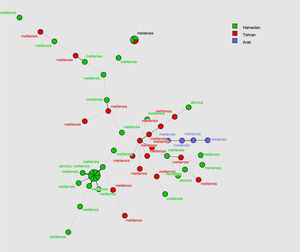
MLVA-16 genotyping resultsThe complete MLVA16 assay was performed targeting panel 1, 2A and 2B loci. The repeat unit size of panel 1 loci was ≥ 9 bp, while that of panel 2A and 2B was up to 8 bp. The PCR products for 16 loci were converted to copy number of TRs. Based on the published data, the polymorphism of each 16 VNTR loci was analyzed by the MLVA-16 method. Based on this method 54 isolates with genetic similarity coefficient of 80% were divided into 46 genotypes. Of those, 22 genotypes were singletons, while 4, 2, 1, and 2 genotypes were represented by 2, 3, 4, and 7 isolates, respectively (Fig. 1). The most prevalent genotype was revealed in 14 strains. There were two more frequent genotypes, each observed in 7 isolates, one genotype was restricted to one geographic region (Hamadan), whereas the other was present in two regions (Hamadan and Tehran). Genotypes present in two or three isolates were from Tehran and Hamadan provinces (Fig. 2). Discriminatory power was determined for each studied locus in this study and according to the results, panel 2B showed the highest discriminatory power [ Bruce-04 (0.837), Bruce-30 (0.806), Bruce-09 (0.787), Bruce-07 (0.772) and Bruce16-(0.766)] while panel 2A displayed a moderate variability [Bruce-18 (0.805), Bruce-19 (0.568), and Bruce-21 (0.358)], and panel 1 demonstrated a limited diversity [Bruce-06 (0.204), Bruce-08 (0.654), Bruce-11 (0.673), Bruce-12 (0.598), Bruce-42 (0.408), Bruce-43 (0.541), Bruce-45 (0.713), Bruce-55 (0.730)].
In the current study a total of 54 Brucella isolates were collected from human and animal samples from three regions of Iran and MLVA-16 was used to assess the genetic diversity among the isolates. Thirty-six (66.66%) isolates were from humans and 18 (33.33%) from animals. Three isolates (5.55%) were identified as Brucella abortus and the remaing 51 were Brucella melitensis. Brucellosis is an atypical zoonotic disease and constant monitoring of animals will largely contribute to the control human brucellosis.14,15 Human brucellosis is most often linked with animal husbandry or consuming unpasteurized milk.16–18 MLVA-16 is a suitable typing method for establishing an epidemiological relationship between isolates in an outbreak to avoid excessive classical epidemiological investigations.19,20 MLVA-16 with a genetic similarity coefficient of 0.8, yielded a total of 46 genotypes, among which 22 were singletons. The frequency of different MLVA genotypes varied in the three studied regions. Three B. melitensis isolates collected from two sheep samples (blood and spleen), one isolates from cattle (liver) and one from human (CSF) in Arak were distinctive in genotypes. Two B. abortus isolated from human blood and one from cattle blood was also distinctive in genotypes. The results of this study showed no correlation between the genotypes of B. abortus isolated from cattle and those isolated from men.
Fifty B. melitensis isolates from human blood in Hamadan shared the same genotype as B. melitensis isolates from sheep blood and B. abortus isolates from cattle spleen in Hamadan, suggesting a possible epidemiological connection among them. These results were consistent to the study of Foster et al. who reported the spread of Brucella from animals to other animals or humans, and the epidemiologic relationship among them.21 Four other genotypes isolated from human blood in Hamedan were very closely related and differed by a single repeat unit at one or two of the most variant loci. Variety in single or double loci may reflect the microevolution of B. melitensis isolates. Additionally, three B. melitensis strains isolated from the blood of three human patients in Hamadan and Tehran, two B. melitensis strains isolated from the blood of animals, two isolates from blood of a human patient and one B. melitensis strain isolated from the liver of sheep were completely matched. These results suggest transmission of Brucella from animals to humans. There are some control measurements and financial compensation for the slaughtered seropositive cattle. However, no such measurement exists for sheep or goat, and therefore, sheep infected with Brucella are one of the main sources for human and animal brucellosis in Iran. This finding is similar to that reported in study from Hai Jiang, China22,23. Similar to previous studies, the highest diversity discriminatory power was found for panel 2B (0.7936) followed by panel 2A (0.557) and panel 1 (0.4760). According to these results, the loci of panel 1 including minisatellite loci with repeat unit length above 9 bp are more conserved than those in panel 2A and panel 2B with more heterogeneous microsatellite loci. The highest discriminatory power was indicated for Bruce30 (0.806) of panel 2B followed by Bruce18 (0.805) of panel 2A.
ConclusionThe MLVA16 analysis on 54 Brucella isolates showed high polymorphism of genotypes. Only two genotypes, each observed in seven isolates, were related to one another and only one of these genotypes belonged to two separate regions.
Availability of data and materialsData are available upon request from corresponding author.
Authors' contributionsSM initiated and supervised the study. SM and FM designed the experiments. FM and AN conducted the experiments. AN conducted the analysis. SM, FM and AN has written the manuscript. All authors have read and approved the manuscript.
FundingThere is no funding for the present study.
Ethics approval and consent to participateThis study was carried according to approved protocol by Ethics committees of Iran University of medical sciences.
Consent for publicationNot applicable.
Competing interestsThe authors declare that they have no competing interests.
Not applicable.