Nontyphoidal Salmonella serotypes are the main cause of human food-borne infection, including several hospitalization cases in the developing countries.
AimTo detect the main serotypes and to characterize the antibiotic resistance of human non-enteric and enteric nontyphoidal Salmonella from clinical isolates in Brazil.
MethodsSalmonella serotypes were identified by microbiological and molecular methods. Susceptibility testing to antibiotics was performed by agar disk diffusion. Real-time PCRs were carried out for the detection of the genus Salmonella as well as serotypes Typhimurium and Enteritidis.
ResultsA total of 307 nontyphoidal Salmonella were isolated from 289 different patients in a reference laboratory (LACEN-RS) from Southern Brazil in a six-year period (2010–2015). There were 45 isolates from emerging cases and 244 from sporadic cases in hospitalized patients. Non-enteric isolates were detected in 42.6% of the patients from sources such as urine, blood and other clinical fluids. Serological and PCR-specific tests demonstrated that Typhimurium (48.4%) and Enteritidis (18.3%) were the most frequent serotypes. Typhimurium isolates were generally resistant to three or more antibiotic classes, while Enteritidis isolates to one or two classes. Typhimurium was the most frequent serotype in all samples (48.4%), mainly among the hospitalized patients (55.6%), and presented the highest rates of multidrug resistance (59.3% of the isolates of this serotype). Further, the prevalence of this serotype increased along the years of the study in comparison to other nontyphoidal Salmonella serotypes.
ConclusionGreater public health attention should be given to prevent salmonellosis in the community and in hospital settings to reduce the rates of Typhimurium strains with multidrug resistance.
Human salmonellosis is one of the most frequently reported foodborne infection and responsible for several community outbreaks.1 Despite global morbidity, mortality is primarily restricted to the developing world.2–4 In Brazil, salmonellosis is considered the most common foodborne bacterial infection with several hospitalizations cases.5
Typhoidal Salmonella (TS) serotypes (Typhi, Paratyphi A and B) are restricted to humans and cause 13.5 million annual episodes of typhoid fever, especially in low- and middle-income countries.6 However, nontyphoidal Salmonella (NTS) infections represent the most burden of salmonellosis worldwide in the last decades.7 Around 80% of the cases are estimated to be caused by NTS serotypes.8 NTS infection is usually self-limited to a gastroenteritis, but some invasive strains can enter the bloodstream and result in a severe life-threatening sepsis, a common cause of bacteremia in both children and adults. Invasive NTS (iNTS) are more severe in the extremes of age and in patients with malnutrition, malaria or HIV infection.9 In Brazil, more than 95% of the salmonellosis cases are caused by NTS.10
Globally, serotypes Typhimurium and Enteritidis are the most common agents of NTS outbreaks.9 In Brazil, Typhimurium has been reported as the main serotype isolated from human systemic infections, while Enteritidis is frequently related to salmonellosis food outbreaks.11,12 Further, some variants lacking flagellar H antigens of these serotypes (mainly Typhimurium) have also been associated with human infections.13
Salmonellosis treatment is usually not required for straightforward gastroenteritis, but it is necessary for an iNTS infection. Chloramphenicol, ampicillin and trimethoprim/sulphamethoxazole were the drugs of choice for many years, but microbial resistance to them have already spread worldwide. Fluoroquinolones (such as ciprofloxacin and ofloxacin) and extended spectrum cephalosporins (such as ceftriaxone and cefotaxime) have been preferred by the clinicians in the last years, although most iNTS have acquired resistance to these both antibiotics too.12,14 This scenario is even worse in regions with high dissemination of multidrug resistance (MDR) Salmonella strains, associated with long period of hospitalization, prolonged illness, high risk of invasive infection and morbidity as well as the more severe outcomes (including death). In recent years, an increasing proportion of MDR Salmonella has been demonstrated in human and animals worldwide.15–17
This study describes the serotype distribution and antimicrobial resistance of NTS isolated from enteric and non-enteric human samples in southern Brazil in a six-year period (2010–2015). In addition, antimicrobial resistance patterns were also investigated in most NTS isolates.
MethodsBacterial isolatesA total of 307 Salmonella isolates were obtained from a laboratory-based surveillance network for enteric pathogens coordinated by the Bacteriology Division from the Central Public Laboratory of the Rio Grande do Sul State (Laboratório Central do Estado do Rio Grande do Sul, LACEN-RS). All these samples were originated from clinical samples collected from cases of salmonellosis in hospitalized patients or ongoing emerging cases in the southernmost Brazilian State, Rio Grande do Sul. Epidemiological data (collection date/season of the year, local/city of collection, clinical sample) were also gathered from the 13 regional clinical laboratories (Coordenadorias Regionais de Saúde – CRSs) of the complete network in Rio Grande do Sul state (with 18 laboratories) and provided by LACEN-RS.
Salmonella identification, serotyping and antimicrobial resistance testingClinical samples were first plated on Salmonella/Shigella solid media (Probac, São Paulo, Brazil). Presumptive Salmonella colonies were then cultured in triple sugar iron (TSI) (PlastLabor, Rio de Janeiro, Brazil), urea, citrate and lysine iron agar (LIA) slants (Probac, São Paulo, Brazil) with aerobic incubation at 37 °C for 24 h. Salmonella suspect colonies were confirmed by agglutination test with polyvalent sera O and H commercial antigens (Probac, São Paulo, Brazil). Complete antigenic characterization and serotype identification were carried out in the National Reference Laboratory for Cholera and Enteric Diseases, Oswaldo Cruz Institute (FIOCRUZ, Rio de Janeiro, Brazil). The somatic (O) antigens were determined by slide agglutination tests and flagellar (H) antigens were determined using a microplate agglutination technique produced by the laboratory. Serotypes were assigned according to the Kauffmann–White–Le Minor (KWL) scheme.18,19
In vitro susceptibility testing to twelve antibiotics was performed by agar disk diffusion according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI).20 The twelve antibiotics used were from nine different antimicrobial categories: aminoglycosides (streptomycin – Str, gentamicin – Gen), cephalosporins (ceftazidime – Caz, cefoxitin – Cfo), penicillins (ampicillin – Amp), quinolones (nalidixic acid – Nal, ciprofloxacin – Cip), tetracyclines (tetracycline – Tet), phenicols (chloramphenicol – Clo), folate pathway inhibitors (trimethoprim/sulfamethoxazole – Tm/Sut), nitrofurans (nitrofurantoin – Nit) and carbapenems (imipenem – Imp). After 24 h of incubation at 36 °C ± 1 °C, clear zones (circles) were identified in the non-resistant samples. The diameter of the inhibition zone was interpreted as resistant, intermediate and susceptible according to CLSI guidelines.20Escherichia coli ATCC 25922 was used as the control bacteria strain. Isolates were classified as MDR if they were resistant to three or more antimicrobial categories according previously described.21
Real-time polymerase chain reaction for the detection of Salmonella and serotypes Typhimurium and EnteritidisDNA extraction of the bacterial isolates was performed by boiling method as described previously.22 A real-time PCR targeting invA gene was carried out for Salmonella generic detection and a duplex real-time PCR targeting fliA-IS200 and safA genes was used for the specific detection of the serotypes Typhimurium and Enteritidis, respectively. Primers/probes sequences and amplification conditions were previously described.23,24 All experiments were carried out in the StepOnePlus™ Real-Time PCR Systems (Thermo Fisher Scientific, Waltham, Massachusetts, USA) with cycling conditions of 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Negative and positive controls were added in all PCRs.
Statistical analysisData were analyzed using the Statistical Package for Social Sciences (SPSS, version 23.0, Chicago, IL). The Pearson's chi-squared test of independence and/or Fisher's exact test were used to verify possible statistical differences between the qualitative variables. Residual adjustment analysis was used to identify the resistance profile of the serotypes to the antimicrobials evaluated. All statistical tests were two-sided and p values < 0.05 were considered as statistically significant.
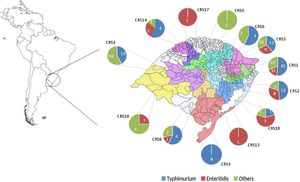
ResultsGeneral epidemiological informationAll the 307 isolates were submitted to the invA real-time PCR and presented positive result to Salmonella. The number of isolates by year was not constant and ranged from 34 in 2010 to 72 in 2012. In addition, there was a similar rate of the Salmonella isolates in Spring (1st September to November 30th) (89, 29%), Summer (1st December to 28th February) (87, 28.3%) and Autumn (1st March to 31th May) (80, 26.1%), but it was observed a reduction of the number of isolates in Winter (1st June to 31th August) (51, 16.6%). All Salmonella isolates were originated from 44 different sub-regions from the Rio Grande do Sul state, covering all geographic macro regions and the most important urban centers in southern Brazil (Fig. 1). The highest number of isolates was obtained in the metropolitan area from the state's capital Porto Alegre (CRS1) with a total of 117 (57.7%) samples, while there was only one isolate in two regions (CR13 and CRS17). No Salmonella isolates were obtained in five macro-regions (CRS7, CRS11, CRS12, CRS15 and CRS16).
The total 307 isolates were from 289 different patients and overall socio-demographic information is demonstrated in Table 1. Salmonellosis frequency was higher in adults with 45 years old or more (105, 36.3%) than in other ages, but there was also an important number of children below five years (54, 18.7%), children and adolescents (34, 11.8%) and adults with less than 45 years old (89, 30.8%). Salmonella was isolated from several different human clinical specimens and classified into non-enteric (blood, urine, abscesses, biological fluids, and aspirates) and enteric (stool) samples according to the source of each sample. Among non-enteric samples 79 (64.2%) were considered invasive because they were isolated from the blood and other clinical fluids such as cerebrospinal, pleural, synovial and ascites (Table 1).
Descriptive analysis of salmonellosis in southern Brazil by year (2010–2015).
| Variables | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | Total |
|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Sex | |||||||
| Male | 15 (45.5) | 23 (56.1) | 28 (43.7) | 31 (63.3) | 35 (53.0) | 15 (41.7) | 147 (50.9) |
| Female | 18 (54.5) | 18 (43.9) | 36 (56.3) | 18 (36.7) | 31 (47.0) | 21 (58.3) | 142 (49.1) |
| Age (years) | |||||||
| 0–5 | 2 (6.1) | 5 (12.2) | 15 (23.1) | 12 (24.5) | 12 (18.2) | 8 (22.9) | 54 (18.7) |
| 6–18 | 2 (6.1) | 7 (17.1) | 6 (9.2) | 9 (18.4) | 6 (9.1) | 4 (11.4) | 34 (11.8) |
| 19–45 | 12 (36.3) | 13 (31.7) | 14 (21.5) | 16 (32.7) | 24 (36.4) | 10 (28.6) | 89 (30.8) |
| ≥45 | 10 (30.3) | 16 (39.0) | 30 (46.2) | 12 (24.5) | 24 (36.4) | 13 (37.1) | 105 (36.3) |
| No data | 7 (21.2) | – | – | – | – | – | 7 (2.4) |
| Origin | |||||||
| Enteric (stool) | 18 (54.5) | 20 (48.8) | 29 (44.6) | 28 (57.1) | 46 (69.7) | 25 (71.4) | 166 (57.4) |
| Non-enteric | 15 (45.5) | 21 (51.2) | 36 (55.4) | 21 (42.9) | 20 (30.3) | 10 (28.6) | 123 (42.6) |
| Clinical samples (non-enteric) | |||||||
| Urine | 3 (21.4) | 4 (20) | 14 (41.2) | 7 (33.3) | 5 (25) | 2 (20) | 35 (29.4) |
| Abscess | – | – | 2 (5.9) | – | 1 (5) | – | 3 (2.5) |
| Aspirate | – | 2 (10) | – | – | – | – | 2 (1.7) |
| Blood | 10 (71.4) | 14 (70) | 17 (50) | 12 (57.2) | 14 (70) | 7 (70) | 74 (62.2) |
| Sterile fluidsa | 1 (7.1) | – | 1 (2.9) | 2 (9.5) | – | 1 (10) | 5 (4.2) |
| Form | |||||||
| Emerging cases | 12 (36.4) | 7 (17.1) | 5 (7.7) | 1 (2.0) | 16 (24.2) | 4 (11.4) | 45 (15.6) |
| Sporadic cases | 21 (63.6) | 34 (82.9) | 60 (92.3) | 48 (98.0) | 50 (75.8) | 31 (88.6) | 244 (84.4) |
A total of 287 isolates from S. enterica subsp. enterica and two isolates from S. enterica subsp. houtenae were detected in the six-year period spanning this study. TS serotypes Typhi and Paratyphi were not detected among the isolates from the subspecies enterica, so all 289 isolates were NTS. The scenario is clearly dominated by serotypes Typhimurium and Enteritidis (193, 66.7%; p < 0.01). Typhimurium was the most frequent one with 140 (48.4%; p < 0.01) isolates, while Enteritidis was detected in 53 (18.3%) of the samples. All these isolates also presented positive result only for the respective serotype in the duplex real-time PCR targeting fliA-IS200 and safA genes.
Among the remaining isolates of the subspecies enterica, 83 (28.7%) were from 21 different serotypes and 11 (3.8%) were not completely serotyped (Table 2). All the 83 isolates of the 21 serotypes and seven samples with incomplete antigenic formula presented negative results in the duplex real-time PCR, but the remaining four isolates presented PCR positive result for serotype Typhimurium (all of them with the antigenic formulae O:4,5). With these four isolates, the prevalence of Typhimurium rises to almost half (144, 49.8%) of all samples. These serotypes were isolated in all regions of the state (Fig. 1).
Salmonella serotypes and antimicrobial resistance level in southern Brazil.
| Serotypes | Isolates | Antimicrobial resistance (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Susceptible | One or two | Three or morea | Missing | ||||||
| n | % | n | % | n | % | n | % | n | |
| Typhimurium | 140 | 48.4 | 10 | 41.7 | 42 | 31.8 | 83 | 72.2 | 5 |
| Enteritidis | 53 | 18.3 | 0 | 0.0 | 39 | 29.5 | 10 | 8.7 | 4 |
| Infantis | 18 | 6.2 | 2 | 8.3 | 12 | 9.1 | 3 | 2.6 | 1 |
| Newport | 13 | 4.5 | 3 | 12.5 | 6 | 4.5 | 3 | 2.6 | 1 |
| S. enterica enterica (others) | 11 | 3.8 | 1 | 4.2 | 4 | 3.0 | 6 | 5.2 | 0 |
| Panama | 10 | 3.4 | 1 | 4.2 | 5 | 3.8 | 2 | 1.7 | 2 |
| Agona | 6 | 2.1 | 0 | 0.0 | 4 | 3.0 | 2 | 1.7 | 0 |
| Give | 4 | 1.4 | 1 | 4.2 | 2 | 1.5 | 1 | 0.9 | 0 |
| Braenderup | 3 | 1.0 | 0 | 0.0 | 3 | 2.3 | 0 | 0.0 | 0 |
| Corvallis | 3 | 1.0 | 2 | 8.3 | 1 | 0.8 | 0 | 0.0 | 0 |
| Derby | 3 | 1.0 | 0 | 0.0 | 3 | 2.3 | 0 | 0.0 | 0 |
| Dublin | 3 | 1.0 | 0 | 0.0 | 2 | 1.5 | 1 | 0.9 | 0 |
| Ohio | 3 | 1.0 | 1 | 4.2 | 1 | 0.8 | 0 | 0.0 | 1 |
| Saintpaul | 3 | 1.0 | 0 | 0.0 | 2 | 1.5 | 0 | 0.0 | 1 |
| Hadar | 2 | 0.7 | 0 | 0.0 | 1 | 0.8 | 1 | 0.9 | 0 |
| Muenchen | 2 | 0.7 | 0 | 0.0 | 2 | 1.5 | 0 | 0.0 | 0 |
| Oranienburg | 2 | 0.7 | 0 | 0.0 | 0 | 0.0 | 1 | 0.9 | 1 |
| Rissen | 2 | 0.7 | 2 | 8.3 | 0 | 0.0 | 0 | 0.0 | 0 |
| S. enterica houtenae | 2 | 0.7 | 0 | 0.0 | 2 | 1.5 | 0 | 0.0 | 0 |
| Albany | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 |
| Bredeney | 1 | 0.4 | 0 | 0.0 | 1 | 0.8 | 0 | 0.0 | 0 |
| Heidelberg | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 |
| Johannesburg | 1 | 0.4 | 1 | 4.2 | 0 | 0.0 | 0 | 0.0 | 0 |
| Mbandaka | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 1 | 0.9 | 1 |
| Ndolo | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 1 | 0.9 | 0 |
| Total | 289 | 100 | 24 | 8.3 | 132 | 45.6 | 115 | 46.1 | 21 |
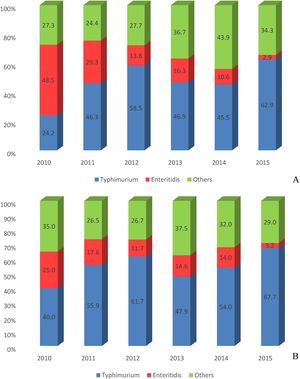
The overall number and frequency of Enteritidis isolates was higher than Typhimurium (and also than the others serotypes) in 2010. The reverse situation (first Typhimurium, second Enteritidis) occurred in 2011. Typhimurium was significantly more frequent than Enteritidis in the other following 4 years (p < 0.01) (Fig. 2A).
Sporadic cases vs emerging casesSalmonella isolates of this study were from sporadic cases in hospitalized patients or associated with ongoing salmonellosis emerging cases. Most isolates (244; 84.4%) were from hospitalized patients and occurred in all geographic macro regions of southern Brazil. Typhimurium was more frequent than all other serotypes, with 135 (55.6%; p < 0.01) isolates, followed by Enteritidis (33; 13.6%). The sum of the other serotypes reached 75 (30.9%) isolates. Interestingly, the overall number and frequency of Typhimurium isolates was much higher than any other serotypes in all years, ranging from 40% in 2010 to 67.7% in 2015 (Fig. 2B).
The remaining 45 isolates were related to 23 regional food-borne salmonellosis emerging cases (9 in 2010, 6 in 2011, 3 in 2012, 4 in 2014 and 1 in 2015). These emerging cases were more frequent in Summer (12, 52.2%), but they were also reported in the three other seasons: Spring (6, 26.1%), Autumn (4, 17.4%) and Winter (1, 4.3%). There were 21 emerging cases with the identification of only one serotype: Enteritidis (14; 60.4%), Typhimurium (3, 14.3%), Infantis (1; 4.8%), Braenderup (1; 4.8%), Corvallis (1; 4.8%) and Johannesburg (1; 4.8%). Noteworthy, there was one important emerging outbreak caused by serotype Infantis with eleven (24.4%) isolates, while all the other ones had only two to four isolates. In one case, two Salmonella serotypes were detected (Agona and Typhimurium).
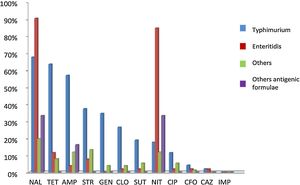
Antimicrobial resistanceA total of 280 (91.4%) NTS isolates were submitted to antimicrobial susceptibility with 12 antibiotics and classified into susceptible and resistant. Sixty-eight (24.3%) were sensitive to all of them, while the other 212 (75.7%) isolates presented resistance to one or more antibiotics. Among these last isolates, 108 (38.6%) were resistant to one or two antimicrobials categories, while the other 104 (37.1%) were resistant to three or more drugs and were considered MDR (Table 2). In an individual evaluation of each antibiotic, it is noteworthy the high frequency of the resistance to Nal (167, 59.2%), Tet (106, 37.6%), Amp (99, 35.1%), Nit (98, 34.2%), Str (92, 32.6%), Gen (54, 19.1%), Cip (44, 15.6%), Clo (43, 15.2%) and Tm/Sut (33, 11.8%). Two other antibiotics showed resistance rates below 5%: Cfo (8, 2.8%) and Caz (4, 1.4%).
In the analysis of the antibiotic resistance according to the serotype, most Typhimurium isolates presented resistance to three or more antibiotics, while the majority of the Enteritidis isolates presented resistance to two antibiotics and most of the other serotypes were susceptible to all tested antibiotics (Table 2). In the analysis of the resistance to different antibiotics categories, Typhimurium was the serotype with the highest number of MDR isolates (83, 72.2%; p < 0.01). Interestingly, twelve (14.4%) Typhimurium isolates presented the multi-resistant profile ACSSut (resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline). In addition, ten (8.7%) Enteritidis isolates and 22 (19.1%) of the other serotypes also were MDR (Table 2). An overall view of the antibiotic resistance according to serotypes (Typhimurium, Enteritidis and others) is demonstrated in Fig. 3.
Antimicrobial resistance rate of different antimicrobial agents in nontyphoidal Salmonella in southern Brazil, 2010 to 2015. The isolates were classified into four serotypes groups: Typhimurium (blue), Enteritidis (red), others (green), and with incomplete antigenic formulae (purple). Antimicrobial resistance was classified according the following antibiotics classes: aminoglycosides (Str, streptomycin, Gen, gentamicin), cephalosporins (Caz, ceftazidime, Cfo, cefoxitin), penicillins (Amp, ampicillin), quinolones (Nal, nalidixic acid, Cip, ciprofloxacin), tetracyclines (Tet, tetracycline), phenicols (Clo, chloramphenicol), folate pathway inhibitors (Tm/Sut, trimethoprim/sulfamethoxazole), nitrofurans (Nit, nitrofurantoin) and carbapenems (Imp, imipenem).
Salmonellosis epidemiology is not well described in South America, unlike in Europe and North America.1,2,8,25 Data from South America continent are scarce because of the lack of an effective tracking system for Salmonella infections and an efficient data recording in the countries. In Africa, for example, there is more data due to the several scientific studies concerning the recent epidemic of iNTS.26,27 Few independent published studies in South America demonstrated a partial landscape in the whole continent, as for example one surveillance study in Colombia, with 4010 Salmonella isolates collected from blood and feces samples over a six-year period, demonstrating the predominance of serotypes Typhimurium (32.5%) and Enteritidis (28.2%).28 In Brazil, wider epidemiological studies date back to more than ten years ago and were performed basically in São Paulo state, located in the southeastern region.10,29
Therefore, the present study aimed at helping to understand the recent epidemiology of this important pathogen in Brazil. The 307 Salmonella isolates were obtained from 289 patients with all ages living in different geographic regions of the southernmost state from Brazil in a six-year period. Interestingly, there was an important frequency of isolates (42.6%) originated from non-enteric human specimens, including 74 (66.7%) blood samples. This result demonstrates the high frequency of occurrence of systemic infections by iNTS strains. The ability of Salmonella to be invasive and cause systemic disease is well recognized and it is estimated that about 5% of the salmonellosis cases result in bacteremia.2,4,30
This scenario is quite different from the main studies in Europe and North America where most isolates are from enteric origin.30,31 Even the previous studies in South America have not demonstrated a so high frequency of non-enteric isolates.28 On oppose, the rates of non-enteric and invasive salmonellosis are larger in Africa than in any other continent. A recent systematic review reported an alarming data of invasive salmonellosis in Africa with one non-enteric to each enteric infection case (ratio of 1:1), contrasting to much lower ratios in Europe is (1:7) and Americas (1:11).26 Despite most samples are from hospitalized patients, it is noteworthy that the results presented here (with a ratio of 1:1.26 in southern Brazil) are very similar to those reported in Africa.26
A more detailed analysis was carried out regarding Salmonella serotypes. The overall results demonstrated the predominance of Typhimurium and Enteritidis serotypes in the clinical cases from southern Brazil. Approximately two-thirds (66.8%) of the Salmonella isolates were from these two serotypes in the whole period of the study. Despite previous studies described the occurrence of these same serotypes in Brazil10,29, it is noteworthy the situation seems to have changed overtime: Typhimurium has a much higher frequency than Enteritidis and other serotypes now. Furthermore, previous studies also presented the occurrence of Salmonella 4,5,12:i:-, a serotype that now is well characterized as monophasic variant of the serotype Typhimurium.10,13,32 Four isolates with a partial antigenic formula presented a PCR positive result to Typhimurium, increasing even more the frequency of this bacteria in the present study (49.8%). There is still a remarkable geographic variation in serotype distribution worldwide, but several other studies demonstrated that Typhimurium displaced Enteritidis as the most frequently isolated serotype.1,10,26 Even in Brazil, recent studies that performed serological and molecular detection demonstrated the high frequency of these two serotypes in other geographical regions.32,33
Salmonella strains of the present study were isolated from emerging or sporadic cases in hospitalized patients. There were 45 isolates from 22 emerging cases caused by nine different serotypes, with the majority of them (13, 59.1%) caused by serotype Enteritidis. Salmonellosis outbreaks are constantly related to foodborne contamination, especially by eating products of animal origin. In addition, Enteritidis has been the most frequently isolated NTS serotype in such outbreaks in southern Brazil.5,11
On oppose, isolates of the hospitalized patients were responsible for the vast majority (84.4%) of salmonellosis cases, with a high frequency of the serotypes Typhimurium (55.6%) and Enteritidis (13.6%). Sporadic cases have been less reported in the scientific literature and usually associated to food origin.1,10–12,34 However, some studies have reported the possibility of nosocomial transmission.35,36 The close proximity of patients to one another, contact by health care workers, and use of shared sanitary facilities favor Salmonella spreading. Nosocomial salmonellosis poses a particular threat to individuals with weakened immune systems who are at higher risk of severe complications requiring antibiotic treatment. It is important to note that serotype Typhimurium was strongly associated to nosocomial transmission in two independent studies.35,36
Antimicrobial resistance was also evaluated and most NTS isolates were resistant to several antibiotics. Nal (59.2%) was the antibiotic most related to cases of resistance as also observed in previous studies in food and human clinical samples.14,29,37 NTS isolates were also resistant to Tet (37.6%), Amp (35.1%) and Nit (34.2%). According to this, in a robust meta-analysis, resistance to sulfonamides (46.4%), Tet (36.9%) and Amp (23.6%) were found in isolates of human origin37. In addition, serotypes Typhimurium (59.3%) and Enteritidis (19.2%) were the most related to the MDR profile. Several data describe that Typhimurium is the main serotype related to MDR and associated with worse prognosis, longer hospitalization time and higher mortality in salmonellosis infections.4,11,18,23,26,36,38 This serotype expresses resistance primarily to quinolones, but it has already been reported to other classes of antibiotics such as penicillins, sulfonamides, tetracyclines and chloramphenicol. Typhimurium isolates with a broad-spectrum resistance to several antibiotics have already been described in Brazil and it is now of great concern for public health.39
Continuous surveillance of Salmonella epidemiology is necessary to monitor trends in serotypes distribution, to prevent spreading of iNTS and to detect drug resistance. This study provided the current serotype prevalence, clinical sources and antibiotic resistance among strains isolated from humans with salmonellosis in southern Brazil. Typhimurium was the most frequent serotype in enteric and non-enteric samples and usually presented MDR. In addition, it was the most frequent serotype detected in hospitalized patients with salmonellosis. New studies are necessary to characterize better the transmission pathways in community and hospitals settings to prevent contamination by this concerning bacteria.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank the Bacteriology Department and others members of LACEN-RS for their collaboration in the development of this study, the staff of the Molecular Diagnostics Laboratory of Lutheran University of Brazil (LDM-ULBRA) who performed technical support and the National Reference Laboratory for Cholera and Enteric Diseases, Oswaldo Cruz Institute (FIOCRUZ, Rio de Janeiro, Brazil) for the complete antigenic characterization and serotype identification. This work was funded by ULBRA and Simbios Biotecnologia. NI and VRL were also financially supported by the National Council for Scientific and Technological Development from Brazil (CNPq – Conselho Nacional de Desenvolvimento Científico e Tecnológico; process numbers 313564/2014-0; 313304/2014-9). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001. RSR, MNS and JMW were further supported by the Coordination for the Improvement of Higher Education Personnel from Brazil (CAPES – Coordenação de Aperfeiçoamento de Pessoal de Nível Superior; process number 181 – 18/12/2012).