This study characterized 30 MRSA isolates from intensive care unit (ICU) environment and equipment surfaces and healthy children. The SCCmec types I, IVa and V were detected in HA-MRSA isolates while CA-MRSA showed the SCCmec type IVa and V. Most isolates were classified as agr group II. All isolates presented the sei gene, and only HA-MRSA were positive for etb e tst genes. Three genotypes were related to Pediatric (ST5/SCCmecIV) and Berlin (ST45/SCCmecIV) clones. The present study showed molecular similarity between CA- and HA-MRSA isolates in hospital and community settings in a Brazilian region.
Staphylococcus aureus is an important human pathogen associated with a broad spectrum of infections in community and hospital settings worldwide.1 Currently, MRSA is the main cause of nosocomial infections in Latin America, and in many Brazilian hospitals the Brazilian Endemic Clone (BEC/ST239/CC8/SCCmecIII) remains the prevalent lineage in these settings.2 However, a number of studies report on a substitution of this well-established clone by the USA400 (ST1/CC1/SCCmecIV) lineage and the Pediatric clone (USA800/ST5/CC5/SCCmecIV).2,3 Meanwhile, the MRSA prevalence in community acquired infections continues to grow. The aim of the present study was to compare molecular characteristics of HA- and CA-MRSA isolates obtained from intensive care units (ICUs) environment and equipment surfaces and from healthy children in public day care centers (DCCs), from Vitória da Conquista, Brazil.
Thirty isolates were identified as MRSA and had their genomic DNA extracted using the PureLink™ Genomic DNA Purification Kit (Invitrogen, Carlsbad, CA, USA). Among them, 19 isolates were collected from ICUs environment and equipment surfaces of two Brazilian public hospitals. The sites, determined by following the routines of each hospital, with a greater possibility of contamination were selected and a total of 117 points were analyzed. S. aureus were isolated from 98 sites and of those 19 (19.28%) isolates were MRSA. One hundred and forty-eight samples of nasal swabs were obtained from healthy children aged one to six years attending DCCs. Children who had received care in hospitals and/or were treated with antibiotics in the last 30 days prior to sample collection were not included. Among 148 children analyzed, S. aureus was isolated in 70 (47.3%). Eleven (15.7%) isolates were MRSA. All institutions are located in Vitória da Conquista, Brazil.
All isolates were characterized for SCCmec types (I–V) and subtype of SCCmecIV (IVa, IVb, IVc and IVd),4agr polymorphism (I–IV),5 and for the presence of 14 virulence genes, including nine enterotoxins (sea, seb, sec, sed, see, seg,seh, sei,sej), two exfoliatins (eta and etb), the toxic shock syndrome toxin (tsst), Panton–Valentine leukocidin (pvl), and icaA.6–9 Moreover, all isolates were analyzed by PFGE after digestion with SmaI (New England Biolabs, Inc., Beverly, MA).10 USA300/ST8/IV, USA400/ST1/IV, USA800/ST5/IV, USA600/ST45/II, USA1100/ST30/IV, and USA100/ST5/II strains were used as standard strains for circulating pandemic clones characterization. One representative isolate of each PFGE type was subjected to MLST and SPA typing techniques.11,12 Statistical analysis: Fisher's exact test was used to compare categorical variables between groups.
The general and molecular characteristics of the 30 MRSA isolates are shown in Table 1. Among them, three SCCmec types were identified. HA-MRSA isolates carried the SCCmec types I (37%; 7/19), IVa (37%; 7/19) and V (26%; 5/19) while CA-MRSA isolates carried SCCmec types IVa (73%; 8/11) and V (27%; 3/11). Moreover, despite that only two agr polymorphisms were found among HA-MRSA strains, agr group II (79%; 15/19) and I (21%; 4/19), three agr were detected in CA-MRSA strains, most isolates related to agr group II (64%; 7/11) (Table 1).
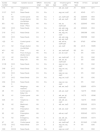
General characteristics of 30 methicillin-resistant Staphylococcus aureus isolates from hospital and community environments in Vitória da Conquista, Brazil.
| Isolate number | Origin | Isolation source | MRSA origin | SCCmec type | agr type | Virulence gene profile | PFGE type | ST/CC | spa type |
|---|---|---|---|---|---|---|---|---|---|
| 17A | H1 | Telephone – ICU | HA | IVa | 1 | etb, sei, icaD | A1 | 45/45 | t371 |
| C137 | DCC 4 | Nasal Swab | CA | IVa | 1 | seg, sei, icaD | A1 | 45/45 | t371 |
| 52 | H2 | Control panel | HA | IVa | 1 | sei, icaA/icaD | A2 | 2228/45 | t004 |
| 59 | H2 | Drug's dilution countertops | HA | IVa | 1 | etb, sei, icaD | A2 | 2228/45 | t004 |
| 85.1 | H2 | Bed-drawer knob – PICU | HA | IVa | 1 | eta, se, icaA/icaD | A2 | 2228/45 | t004 |
| C60 | DCC 2 | Nasal Swab | CA | IVa | 1 | eta, seb, seg, sei, sej, icaD | A2 | 2228/45 | t004 |
| C18 | DCC 1 | Nasal Swab | CA | V | 4 | eta, seg, sei, icaD | J | 398/398 | t242 |
| C83 | DCC 2 | Nasal Swab | CA | V | 4 | eta, seb, seg, sei, sej, icaD | J | 398/398 | t242 |
| 12D | H1 | Control panel – ICU | HA | V | 2 | etb, sei tst, icaA/icaD | B1 | 676/5 | t9784 |
| 47.1 | H2 | Drug's dilution countertops | HA | V | 2 | sei, icaD | B2 | 461/5 | t5085 |
| 43.1 | H2 | Floor | HA | V | 2 | sei, icaA/icaD | B3 | 5/5 | t311 |
| 91 | H2 | Floor of drug's dilution room | HA | IVa | 2 | sei, tst, icaD | C | 5/5 | t242 |
| 101 | H2 | Sink PICU | HA | IVa | 2 | sei, icaA/icaD | C | 5/5 | t242 |
| 27A | H1 | Baby Crib | HA | IVa | 2 | etb, sei, tst, icaA/icaD | C | 5/5 | t242 |
| C138 | DCC 4 | Nasal Swab | CA | IVa | 2 | seg, sei, sej, icaD | C | 5/5 | t242 |
| C48 | DCC 2 | Nasal Swab | CA | IVa | 2 | seg, sei, sej, icaD | C | 5/5 | t242 |
| C54 | DCC 2 | Nasal Swab | CA | IVa | 2 | seg, sei, sej, icaD | C | 5/5 | t242 |
| C77 | DCC 2 | Nasal Swab | CA | IVa | 2 | seg, sei, icaD | C | 5/5 | t242 |
| C94 | DCC 3 | Nasal Swab | CA | IVa | 2 | seg, sei, sej, icaD | C | 5/5 | t242 |
| 16A | H1 | Weighing-machine | HA | V | 2 | etb, sei, icaD | D | 2226/5 | t3374 |
| 28 | H1 | Control panel – Baby Crib | HA | I | 2 | etb, sei, icaD | E | 1427/5 | t5046 |
| 29 | H1 | Control panel ICU | HA | I | 2 | etb, sei, tst, icaA/icaD | E | 1427/5 | t5046 |
| 33A | H1 | Telephone – ICU | HA | I | 2 | etb, etb, sei, icaD | E | 1427/5 | t5046 |
| 39 | H2 | Medicine's fridge | HA | I | 2 | etb, sei, icaD | F | 3050/45 | t3374 |
| 41 | H2 | Prescription paper | HA | I | 2 | etb, sei, icaD | F | 3050/45 | t3374 |
| 62 | H2 | Prescription paper | HA | I | 2 | etb sei, icaD | F | 3050/45 | t3374 |
| 74 | H2 | Emergency stretcher | HA | I | 2 | eta, etb, sei, icaA/icaD | F | 3050/45 | t3374 |
| 67 | H2 | Control panel | HA | V | 2 | sei, icaA/icaD | G | 3019/45 | t11545 |
| C152 | DCC 4 | Nasal Swab | CA | V | 2 | seg, sei, sej, icaD | H | 5/5 | t242 |
| C80 | DCC2 | Nasal Swab | CA | IVa | 2 | seg, sei, icaD | I | 5/5 | t002 |
H1, Hospital Municipal Esaú Matos; H2, Hospital Geral de Vitória de Conquista; DCC, day care center; HA-MRSA, hospital-acquired MRSA; CA-MRSA, community-acquired MRSA; ICU, intensive care unit; PICU, pediatric intensive care unit; SCCmec, Staphylococcal cassete chromosome mec; agr, accessory gene regulator; PFGE, pulsed field gel electrophoresis; ST, sequence type; CC, clonal complex.
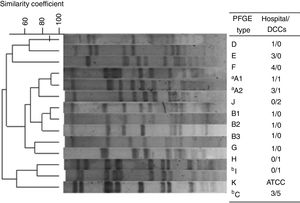
Out of the 30 MRSA isolates, 13 different PFGE patterns were identified grouped into 10 genotypes, from A to J (Fig. 1). Among them, 10 STs were identified as being related to three clonal complexes (CC): 5, 45 and 398. For all 19 HA-MRSA isolates, seven different clones (>80% similarity) were found (A–G) to be related to CC5 (53%; 10/19) and CC45 (47%; 9/19) and nine spa types were characterized. Meanwhile, for CA-MRSA isolates, five lineages were described (A, C, H, I and J) belonging to CC5 (64%; 7/11), CC45 (18%; 2/11) and CC398 (18%; 2/11) as well as four spa types, the most prevalent being t242 (73%, 8/11). Interestingly, two lineages were commonly found among HA- and CA-MRSA isolates (n=14; 47%), sharing the same molecular characteristics, related to A and C genotypes. Overall, the Pediatric Clone (USA800/ST5/SCCmecIV) was related to nine isolates (30%; 9/30), both CA-MRSA (n=6) and HA-MRSA (n=3), eight of them sharing the same PFGE profile (C). Moreover, the STs 45/CC45, characterized as the Berlin Clone (USA600/ST45/SCCmecIV), and 2228/CC45 were found among both groups, related to genotypes A1 (2/30; 6.7%) and A2 (4/30; 13.3%), respectively.
Dendrogram of the PFGE patterns related to 30 CA- and HA-MRSA isolates. Isolates showing a similarity coefficient ≥80% were considered genetically related. DCC, day care centers. One isolate, S. aureus strain ATCC 29213/ST5 was used as control. aRelated to USA600 lineage; bRelated to USA800 lineage.
In regards to virulence genes, the sei gene was present in all 30 MRSA isolates (Table 1). Despite that the seg (100%) and sej (64%; 7/11) genes were found to be associated only with CA-MRSA isolates (p-value<0.05), the etb (63%; 12/19) (p-value<0.05) and tst (21%; 4/19) genes were only detected among HA-MRSA isolates. The eta gene was present in CA-MRSA (27%) and HA-MRSA (10.5%) isolates. The icaD gene was related to both HA- and CA-MRSA, whereas icaA/icaD was detected only among HA-MRSA isolates (p-value<0.05).
The epidemiology of MRSA infections is very dynamic and the substitution of well adapted hospital clones by community ones has been shown by several authors.2,3,13 However, there are few studies comparing HA- and CA-MRSA isolates in Brazilian studies.
A prevalence of SCCmec types IV and V, typically from the community, were reported in all CA-MRSA isolates, and were the main types found among HA-MRSA isolates (63%) as well. The distinction between MRSA isolates from community and healthcare facilities became cloudy with the replacement of HA-MRSA lineages by CA-MRSA in the hospital setting. In a study conducted in Brazil describing the molecular characteristics of isolates collected from healthy children, most isolates were related to SCCmec III and only three isolates showed the SCCmec type IV and one type V.14 On the other hand, Caboclo et al.15 showed dissemination of the USA400/SCCmec IV, a community lineage, at a military hospital in Rio de Janeiro.2 In Brazil, the report of isolates carrying the SCCmec type V is still rare, but in the present study this cassette type was present in both groups evaluated. The presence of both SCCmec types IV and V among healthy and hospitalized people seems to be related to the exposure of healthcare professionals, hospital staff, visitors, and patients to a wide range of pathogens.16
The MLST revealed a wide diversity of STs in both environments. ST5, ST45 and ST2228 (single locus variant ST45) were found among CA-MRSA and HA-MRSA strains. The ST5 isolates has emerged in hospital and community isolates, while ST45 has most often been found in the CA-MRSA carrying SCCmec IV.1,17 The spa typing also showed great genetic variation among the analyzed isolates. Most CA-MRSA isolates shared common spa types with HA-MRSA isolates.
The Clonal complexes – CC5 and CC45 – which represented 100% of HA-MRSA and 82% of CA-MRSA isolates in this study, are among the clonal groups known to be involved in a global pandemic caused by MRSA.1 Although CA-MRSA isolates usually have different molecular characteristics than HA-MRSA, most community isolates in this study shared the same genotype of those from hospitals, mainly in pulsotypes A and C (n=14; 47%). The C and I clones were related to the Pediatric clone (ST5/CC5-SCCmecIV). The A1 and A2 subtypes were related to the Berlin clone (ST45/CC45-SCCmecIV). Thereby, both Pediatric and Berlin clones are present in children attending day care centers (55% and 18%, respectively) and on ICUs environmental surfaces (16% and 21% respectively), sharing the same genetic background. This similarity between genetic groups of CA- and HA-MRSA may indicate an eventual co-transmission between the community and hospital settings.
The Berlin clone (CC45), recently described in the northeast of Brazil among MSSA (2%) isolates from nosocomial infections, has the ability to cause high mortality in patients with bloodstream infections and a large global spread capacity.18 On the other hand, the Pediatric clone (CC5) has emerged in Brazilian hospitals,19 and this is the first detection in healthy children and objects in ICUs and equipment surfaces, suggesting that this clone is spreading from Brazilian hospitals to the community, which can act as a reservoir and contribute to the spread of this pathogen.
The accessory gene regulator group I and II predominated in the community and hospital isolates. These groups have been associated with endocarditis and suppurative infections.20 This study revealed that enterotoxin genes were more prevalent in CA-MRSA isolates, a result also found by Xie et al.17 comparing HA- and CA-MRSA in hospital isolates in China, while the exfoliatin b (etb) gene predominated among HA-MRSA isolates. The presence of superantigen genes in MRSA isolates from hospital surfaces and healthy individuals implies the possibility of increased bacterial dissemination and more severe infections. Genes related to biofilm production were detected in all strains. The finding of this gene is important because infections associated with biofilm production are usually recurrent, aggravate nosocomial infections, and act as a barrier to antimicrobial action.
Our results demonstrated high genetic diversity among MRSA isolates, although most of the isolates are related to the CC5 and CC45 showing the importance of local studies to better understand the dynamics involved in the spread and pathogenicity of MRSA lineages.
In conclusion, the CA- and HA-MRSA isolates sharing similar molecular characteristics irrespective of their environment origin as well as the discovery of international clonal lineages demonstrate the dissemination ability of MRSA and the risk of community and hospital infections. It underscores the need for public health officials to monitor these populations, sites and to develop strategies to reduce the prevalence of these MRSA clones on hospital surfaces and DCCs. These environments could act as important reservoirs for future community and hospital infections.
Conflicts of interestThe authors declare no conflicts of interest.
We thank the Hospitals and DCCs, children and parents/guardians for allowing this research to be conducted, and AcademicEnglishSolutions.com for proofreading. This study was supported by Programa de apoio a pesquisadores emergentes da UFBA (PRODOC 02/2011).