This study was conducted to provide information on the genetic diversity of human parvovirus B19 (B19V) circulating in the municipality of Niterói, Rio de Janeiro, Southeast Brazil during 1996–2006, a period with two distinct outbreaks of B19V infection: 1999–2000 and 2004–2005. A total of 27 sera from patients with erythema infectiosum and five sera from HIV-infected patients that tested positive for B19V DNA during the study period were analyzed. To genotype B19V strains, a semi-nested PCR for partial amplification of the capsid gene was performed and sequence analysis revealed that 31 sequences belonged to subgenotype 1a (G1a) of the main genotype 1 and one sequence was characterized as subgenotype 3b (G3b). The phylogenetic tree supported the division of the G1a into two well-defined clades with 1.3% of divergence. The low diversity of the G1a strains may be explained by the fact that all patients had acute B19V infection and 30/32 sera were collected during two distinct outbreaks. The G3b strain was from an HIV-infected patient who seroconverted to anti-B19 IgG antibodies in September/2005. This is the first report of G3b in the state of Rio de Janeiro.
The human parvovirus B19 (B19V), a member of the Erythroparvovirus genus (Parvoviridae family), is a small, non-enveloped icosahedral virus composed of two structural proteins VP1 (83kDa) and VP2 (58kDa) surrounding a linear single-stranded DNA genome of approximately 5.6Kb.1,2 Currently, three distinct genotypes of B19V have been identified: (i) genotype 1, with subtypes 1a (the prototypic virus) and 1b; (ii) genotype 2 (A6 and LaLi strains); and (iii) genotype 3, with subtypes 3a (V9 strain) and 3b (D91.1 strain).2–4 Genotype 1 is the most prevalent worldwide, genotype 2 has been detected in patients born before 1973 from European countries, the United States and Brazil, and genotype 3 predominates in parts of West Africa.2,5
The most common clinical presentation of B19V infection in immunocompetent individuals is the erythema infectiosum (EI) or fifth disease, a self-limited childhood exanthema characterized by a “slapped-cheek” rash. Adults with EI, particularly women, frequently present clinically with arthropathy. It is assumed that EI and the joint symptoms are the result of deposition of immune complexes.2,6,7
Previous studies conducted by our group demonstrated that B19V infection is a common event in schoolchildren aged five to nine years old and that EI outbreaks seem to occur every 4–5 years.6,7 All three genotypes of B19V have been reported in patients presenting with acute and/or persistent B19V infection in Brazil.5,8–12 Nonetheless, there is a lack of information regarding the genotypes of B19V circulating during outbreaks of EI in our country.
This study was conducted to provide information on the genetic diversity of B19V circulating during a period spanning two outbreaks of EI in the municipality of Niterói, Rio de Janeiro state. In addition, B19V strains from HIV-positive patients infected during the study period were also characterized.
The serum samples analyzed in this study were collected during a 10-year-period (1996–2006) from 131 patients with exanthematic disease, 28 of them with arthropathy, attending the Infectious Diseases Department, Antonio Pedro University Hospital, Federal Fluminense University in Niterói, RJ. These sera had previously tested positive for anti-B19V IgM using a commercial enzyme immunoassay (Biotrin International, Dublin, Ireland).6,7 Of 131 patients, 79 were female. Their ages ranged from 9 months to 54 years. In addition, sera from five HIV-infected adult patients who attended the general medical outpatient clinic for routine care and were found to be positive for B19V DNA during the study period were also examined. These cases were retrieved from a study to estimate the frequency of B19V seroconversion in a cohort of HIV-infected patients.13 Written informed consent was obtained from all participants and the project was approved by the Hospital Review Board (CMM/HUAP 134/05).
For molecular detection of B19V and sequencing, viral DNA was extracted from serum samples using a QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). Genotyping of B19V strains was performed with a semi-nested PCR using the primer pairs P12/P16 (4127–4689) and P13/P16 (4214–4689) for partial amplification of the VP1/VP2 capsid gene.5 The 476-bp amplicons were purified using the illustra GFX™ PCR DNA and Gel Band Purification kit (GE, Healthcare®, Buckinghamshire, UK) and then subjected to direct sequencing using the BigDye terminator v. 1.1 cycle sequencing kit (Applied Biosystems, CA, USA) and an ABI Prism® 3730 DNA analyzer (Applied Biosystems, CA, USA). Both strands of each amplicon were sequenced at least twice. The nucleotide sequences obtained during this study were deposited in the GenBank database under accession numbers KP115293–KP115325.
For sequence analysis and phylogeny, sequences were aligned using the BioEdit Sequence Alignment Editor v7.2.5. Phylogenetic trees were constructed by Bayesian Markov Chain Monte Carlo (MCMC) statistical framework implemented in the BEAST v1.7.414 and maximum likelihood (ML) available in the MEGA v.6.0 software.15 The evolutionary model Hasegawa-Kishino-Yano plus gamma (HKY+γ) was the substitution model used as the best fitted by Model test 3.7.16
To determine the selection pressure acting at variable positions we used the Fixed effects likelihood (FEL), Random effect likelihood (REL) and Single likelihood ancestor counting (SLAC) of the Datamonkey software.17
B19V infection was characterized by variable combinations of fever, flu-like symptoms, arthropathy, and gastrointestinal symptoms. All cases presented with an erythematous maculopapular rash. “Slappedcheek” appearance and reticular or lace-like rash were seen in only 30% of the children. No adult presented this typical rash. Acute arthropathy was reported more frequently in adults, mainly in women, and in general it was symmetrical, affecting preferentially the small joints of the hands, feet, knees and wrists. In most of the cases polyarthropathy completely resolved within two weeks. No clinical manifestations of B19V infection were detected in the medical records of the five HIV-infected adult patients during the seroconversion period.
A 476bp fragment of the B19V capsid gene was amplified by PCR from 67/131 (51.1%) sera from patients with EI. The majority of the positive samples were collected during the 1999–2000 (n=40) and 2004–2005 (n=23).
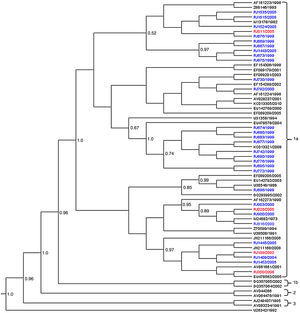
Nucleotide sequences of 27/67 amplified B19V DNA had a satisfactory quality for analysis. All the 27 sequences detected in EI patients belonged to subgenotype 1a (G1a) (Fig. 1).
Phylogenetic analysis of B19V VP1/VP2 partial sequences (427bp), obtained from patients with erythema infectiosum (blue) and HIV-infected patients (red) in Niterói, RJ, Brazil, from 1996 to 2006. The reference sequences are showed with their GenBank accession numbers in black. The Bayesian Maximum Clade Credibility tree was based on the relaxed molecular clock and the MCMC analysis was run for 1×109 generations to achieve the convergence of parameters.
Amongst the five B19V sequences obtained from the HIV-infected patients, four (RJ109/2002, RJ011/2005, RJ026/2005, RJ350/2006) were grouped into subgenotype G1a and the fifth sequence (RJ071/2005) into 3b (G3b). Of these five serum samples, three were collected during the 2004–2005 outbreak.
The phylogenetic tree supported the division of the G1a in two well-defined clades (posterior probability=1). The sequences from EI patients were distributed along the two clades while 3/4 sequences from HIV-infected patients were kept on the same clade (Fig. 1).
The average nucleotide distance observed between the G1a sequences of this study with other G1a and G1b was 1.6% and 5.6%, respectively, and with G3b was 14.5%. Within subgenotype 1a, the viral sequences were more closely related to each other (d=1.0%) than to G1a isolates from Brazil or other countries (d=1.2–1.8%). The nucleotide divergence between the two G1a clades was 1.3% (amino acid divergence of 0.1%) and the genetic distance within the clades was very low (p-distance=0.9% and 0.6%).
Comparison of the 427bp fragment with G1a sequences retrieved from GenBank revealed four nucleotide substitutions at positions 4260 (A→T), 4267 (A→C), 4262 (A→T), and 4552(T→C) that were identified only in strains of this study during 1999–2000 outbreak (RJ674/1999, RJ676/1999, RJ677/1999, RJ680/1999, RJ683/1999, RJ690/1999, RJ695/1999, RJ/742/1999, RJ776/1999, RJ772/1999, RJ903/2000). Another four changes at positions 4349 (C→T), 4414 (T→C), 4507(A→G) and 4570 (C→T) were also found in Brazilian strains from Pará and São Paulo. Among the eight nucleotide changes all but one were synonymous (position 4260), a finding easily explained since 6/8 changes occurred in the 3rd codon position.
The examination of the selective pressures by the FEL, REL and SLAC algorithms of Datamonkey package demonstrated that the region is subject to strong negative selection. No positively select site was found.
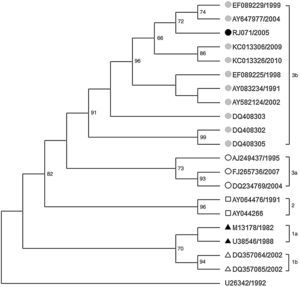
One sequence (RJ071/2005) from a serum of an adult female HIV-infected patient receiving antiretroviral therapy who seroconverted to anti-B19 IgG antibodies in September 2005 showed more than 14.5% of divergence with subgenotype 1a in the 427bp VP1/VP2 region, 12.2% with genotype 2 and 1.8% with subgenotype 3b. The phylogenetic tree based on these data showed that this sequence clustered in the same branch as others belonging to genotype 3b. It shared higher nucleotide homology (99.8%) with other Brazilian 3b sequences obtained from a patient with systemic lupus erythematous from Pará (EF0892229/1999) and another with anemia after a living-donor kidney transplant from São Paulo (AY647977/2004) (Fig. 2).5,9
Maximum likelihood phylogenetic tree of 427bp fragment of the VP1/VP2 capsid gene of B19V (Genotype 3b) based on Hasegawa-Kishino-Yanoda plus gamma (HKY+γ) evolution model. Bootstraps values (>50%) based on 2000 replicates are shown above the branches. The reference sequences are showed with their GenBank accession numbers.
This analysis of B19V genotypes circulating from 1996 to 2006 was performed since there was a lack of information regarding the genotypes of B19V circulating during outbreaks of EI in Brazil. During the study period two distinct outbreaks of B19V infection occurred: 1999–2000 and 2004–2005.6,7 We demonstrated the predominance of subgenotype 1a. This is consistent with previous studies performed in Brazil (North, Midwest, South, and Southeast Brazil) in which the majority of the sequences from sera of patients with acute B19V infection belonged to subgenotype 1a.5,9,10,12 Sequences from subgenotype 1a of this study had average genetic diversity of 1.0% and the diversity from 3b isolates was around 14.5%. These results are consistent with previous studies in which the same region of VP1/VP2 gene was analyzed.3,4,5
The first evidence that two groups of subgenotype 1a coexist around the world arose from the phylogenetic analysis of strains from Dutch blood donors and patients with fifth disease.18 The subdivision of subgenotype 1a into two groups was also proposed by Freitas et al.5 in a study that analyzed the same 427bp fragment of VP1/VP2 region from patients with several clinical manifestations of B19V infection in the Amazon region (North Brazil). Recently, Slavov et al.12 examining a 446bp fragment corresponding to the NS-1 gene of B19V from patients with hemoglobinopathies and blood donors in the State of São Paulo (Southeast Brazil) also reported the division of subgenotype 1a into two clades.
In agreement with previous reports, these results show two well-defined clades into subgenotype 1a. The genetic distance between the two clades was very low (d=0.013) which is consistent with other studies.5,18
The low diversity and occurrence of non-synonymous substitutions of the strains G1a of this study may be explained by: (i) all sera were from patients with acute infection6,7,13; (ii) the majority of the sera were collected during two distinct outbreaks; and (iii) almost all the nucleotide substitutions occurred in the 3rd codon position which may not lead to amino acid change.
Nucleotide changes might suggest temporal or geographic clustering but neither the genotypic grouping nor the co-circulation of multiple lineages of G1a could be associated with a particular outbreak. It is important to note that the amplified fragment correspond to the variable carboxy-terminal region of VP1/VP2 capsid gene, a region that contains several neutralizing epitopes.1
Unlike acute infection, B19V isolates from patients with persistent infection exhibit a higher degree of variability, both at DNA and protein level, due to a low but continuous virus replication over prolonged periods of time which may contribute to accumulated mutations.1
The association of high nucleotide diversity with low amino acid variability observed in B19V is consistent with the apparent lack of difference in clinical manifestations and antigenic reactivity in serological tests.1,3
In Brazil, all three B19V genotypes have been identified. Whilst genotypes 1 and 3 have been detected in serum samples from patients with EI as well in sera and bone marrow of patients with B19-related hematological symptoms,5,8–12 genotype 2 has only been detected in the bone marrow of older patients (mean age 62.5 years) with cytopenias of unknown origin.11,19
Surprisingly, a sequence obtained from a serum of an HIV-infected patient during 2005 oubreak20 was characterized as subgenotype 3b. The origin of this subgenotype is difficult to determine since the serum was collected during routine follow-up examination of the patient under antiretroviral therapy. No travel information was available on medical records. Furthermore, this is the first report of genotype 3b in the state of Rio de Janeiro, Southeast Brazil.
The predominance of genotype 1 (B19V), the low frequency of genotype 3 and the absence of genotype 2 in our study are similar to those previously found in other regions of Brazil.5,8,9,11,12,19 As the PCR assay used amplifies all three genotypes of B19V, the lack of detection indicates the absence of genotype 2 in these patients. Genotype 2 DNA has been found mostly in tissues of individuals born before 1960.2,19 Most sequences analyzed in this study were from acute infections in patients less than 40 years old.
The current study represents a further contribution to the molecular characterization of B19V in Brazil since it presents an analysis of the genotypes circulating during two distinct outbreaks of erythema infectiosum in our country.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Universidade Federal Fluminense – Pró-Reitoria de Pesquisa, Pós-Graduação e Inovação (UFF-PROPPI), Fundação de Amparo à Pesquisa Carlos Chagas Filho (FAPERJ) and Instituto Oswaldo Cruz – Fundação Oswaldo Cruz (IOC-FIOCRUZ). The authors wish to thank Prof. Felipe Piedade Gonçalves Neves and André Victor Barbosa for technical support on behalf of Plataforma de Sequenciamento de DNA, Laboratório Multiusuários de Microbiologia e Parasitologia (LMMP) of Universidade Federal Fluminense.