The goal of this study was to investigate the prevalence of occult HBV infection in a reference center for the Northern Brazil from 2005 to 2015 and to identify mutations associated with occult hepatitis B. Molecular analysis was performed on 110 serum samples in which anti-HBc was the only positive serological marker. Regions of the HBV genome were amplified by polymerase chain reaction to detect HBV DNA. A prevalence of 4.1% (793/18,889) for anti-HBc alone was identified. Molecular analysis revealed a prevalence of occult HBV infection of 0.04%. HBV DNA detected were identified in individuals who underwent hemodialysis, infected with the hepatitis C virus and from area of high endemicity for HBV. Direct DNA nucleotide sequencing and phylogenetic analysis identified that genotypes A and D and mutations E164D, I195M, P217L and P120S were associated with occult HBV infection in the S gene. This study contributed with epidemiological and molecular information on Northern Brazil samples with a suggestive profile of occult HBV infection in addition to reinforcing the importance of molecular diagnosis in this type of infection.
Approximately one-third of the world's population has serological evidence of current or previous hepatitis B virus (HBV) infection, and approximately 350-400 million individuals are chronic carriers of the virus.1,2 Chronic HBV infection is a dynamic process.2 The spectrum of the disease and the natural history of chronic HBV infection are diverse and heterogeneous, ranging from inactive carrier state to progressive chronic hepatitis B, which can progress to cirrhosis and hepatocellular carcinoma.2
HBV is an enveloped DNA virus belonging to the family Hepadnaviridae, genus Orthohepadnavirus, species Hepatitis B virus.3 The viral genome is circular, and the partially double-stranded DNA encompasses approximately 3,200 base pairs divided into four partially overlapping genes: preS/S, preCore/Core, Pol and X3. These genes encode seven different proteins: three surface proteins (small, medium and large S), two core proteins (core antigen (HBcAg) and e-antigen (HBeAg)), viral polymerase and regulatory X protein.3 Ten genotypes (A-J) have been described and are divided into several subgenotypes and subtypes, with different ethnic-geographical distributions among genotypes.4,5
Several definitions of occult HBV infection (OBI) or occult hepatitis B have been suggested. In 2008, the European Association for the Study of the Liver defined OBI as the presence of HBV DNA in the liver (with or without the presence of HBV DNA in the serum) of hepatitis B surface antigen (HBsAg)-negative individuals tested with routine laboratory assays.6 In these individuals, HBV DNA in the serum can be detected or undetected, and when it is detected, levels are usually very low (< 200 IU/mL).6 The prevalence of OBI ranges from 1% to 95% in different parts of the world. These prevalence rates are influenced by several factors, such as 1) geographic differences (endemicity); 2) different patient characteristics (risk groups); and 3) differences in diagnostic techniques used (different sensitivities).6,7
OBI may have clinical impacts, such as the possibility of virus transmission (mainly through blood transfusion and liver transplantation), risk of reactivation of the disease, contribution to the progression of liver disease, and development of hepatocellular carcinoma.8-10 Due to these possible clinical implications, there is currently an increase in the number of studies involving OBI, especially with respect to its prevalence and the molecular mechanisms involved in this type of infection.
Detection rates for hepatitis B in Brazil showed little variation in the last 10 years, with slight downward trend from 2015, reaching 6.6 cases per 100,000 inhabitants in 2019.11 From 2009 to 2019, detection rates in the South, North and Center-West regions were higher than the national rate, while the lowest rates were observed in the Northeast region.11 Information about the situation regarding OBI in Northern Brazil is still scarce due to lack of molecular studies investigating this type of infection. The present study aimed to determine the prevalence of OBI at a reference center in Northern Brazil for the diagnosis of viral hepatitis and to identify the mutations involved in the nondetection of the virus by commercial serological tests.
Materials and methodsSamplemThe present descriptive and cross-sectional study was approved by the Research Ethics Committee of Evandro Chagas Institute (approval number 1,476,122). The selection of samples for the study was based on a retrospective survey conducted through an active search of the Database of the Hepatology Department of the Evandro Chagas Institute (IEC, for its initials in Portuguese). The IEC is located in the city of Belém, state of Pará and is considered a regional reference center (Northern Brazil) in the diagnosis of viral hepatitis. In the period from 2005 to 2015 a total of 18,889 serum samples were evaluated by enzyme immunoassays, and the selected samples were those that were reagent (positive) to total anti-HBc but non-reagent (negative) to HBsAg and anti-HBs and were screened for HBV-DNA to identify the presence of occult HBV infection. All serological tests for hepatitis B (HBsAg, anti-HBs and total anti-HBc) were based on enzyme-linked immunosorbent assay (ELISA) using commercial kits commonly employed in routine clinical laboratory testing. According to availability, storage conditions and serum volume, a total of 110 samples with suggestive serological profile (anti-HBc alone) of occult HBV infection were analyzed by molecular biology tests.
The molecular study for the detection of HBV DNA was developed using previously extracted and isolated samples of viral DNA, which were stored under freezing conditions (-20°C). Viral DNA was extracted, stored and identified according to the standard procedure of the molecular biology laboratory of the IECʼs Hepatology Department. DNA extraction was performed in an automated manner according to the manufacturer's instructions (Abbott, USA), using magnetic microparticles to isolate the nucleic acid.
PCR detection of HBV DNAMolecular analysis was performed using in-house nested PCR, as previously described.12,13 Surface antigen (S), polymerase (Pol) and core (C) partial regions of the HBV genome were amplified by nested PCR (S-Pol region and C region) to detect HBV DNA. The S-Pol region was amplified by nested PCR using specific primers, yielding a fragment of 734 base pairs (bp). The amplification conditions were as follows: initial denaturation at 94°C for 1 min; followed by 34 cycles at 94°C for 30 sec, 56°C (first PCR)/50°C (second PCR) for 30 sec and 72°C for 30 sec; and a final extension step at 72°C for 7 min.
For the amplification of the C region, specific primers were used, yielding a fragment of 248 bp. For the amplification reaction, the following conditions were used: initial denaturation at 94°C for 2 min; followed by 25 cycles at 94°C for 1 min, 42°C (first PCR)/50°C (second PCR) for 30 sec and 72°C for 1 min; and a final extension step at 72°C for 7 min. The PCR products were separated by horizontal electrophoresis on a 1% agarose gel with SYBR Safe DNA gel stain (Invitrogen, USA) and observed under ultraviolet light.
Nucleotide sequencing and phylogenetic analysisSamples with HBV DNA detected by nested PCR were subjected to direct DNA nucleotide sequencing to identify the viral genotype and mutations associated with occult HBV infection. The PCR products were sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), following the manufacturer's instructions. Direct DNA sequencing was performed with an ABI 3500 genetic analyzer (Applied Biosystems, USA). The DNA sequences obtained for the S-Pol region were edited and aligned with the Geneious software version 9.1.5, along with sequences from the different HBV genotypes and subgenotypes available in GenBank. Subsequently, phylogenetic trees were constructed using the maximum likelihood method in the Geneious software. Bootstrap analysis with 1,000 replications was performed to evaluate the reliability of the branches (values ≥ 70) of the phylogenetic tree. For the identification of mutations that escaped detection by enzyme immunoassays, amino acids were deduced from the sequencing of nucleotides in the S-Pol region using Geneious software version 9.1.5. The sequences obtained were aligned with wild-type HBV sequences and visually analyzed to detect S gene mutations. Mutations in the gene encoding the viral polymerase were also investigated.
ResultsA total of 18,889 serum samples were selected from individuals who underwent serological tests for hepatitis B in the Hepatology Department of the Evandro Chagas Institute from January 2005 to December 2015. A prevalence of 4.1% (793/18,889) for anti-HBc alone was identified during this period. A total of 110 samples with anti-HBc alone were selected. These samples were tested for the presence of viral nucleic acid, and HBV DNA was detected in 8.1% (9/110), indicating an occult HBV infection prevalence of 0.04% (9/18,889). Samples with HBV DNA detected were identified in individuals who underwent hemodialysis (two samples), infected with the hepatitis C virus (HCV) (six samples), and from an area of high endemicity for HBV (one sample).
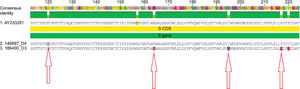
The samples with HBV DNA were subsequently subjected to direct DNA sequencing to identify the viral genotype and the presence of mutations associated with occult HBV infection. Phylogenetic analysis identified genotypes A (subgenotype A1) and D (subgenotypes D3 and D4) (Table 1), and the alignment of the amino acid sequence of the S region revealed four different mutations associated with occult HBV infection: E164D, I195M, P217L and P120S (Fig. 1). In the Pol region, no mutations associated with occult HBV infection were identified.
Identification of HBV genotype in samples with anti-HBc alone and HBV DNA detected in Northern Brazil.
In samples 3,4,7,8 and 9 it was not possible to identify the HBV genotype. PA: state of Pará/Brazil.
The global prevalence of OBI is estimated to be between 4 and 25% among individuals who are positive for anti-HBc alone.8,10,14 In the present study, all samples analyzed had anti-HBc as the only positive serological marker, and the prevalence of occult HBV infection was 0.04%.
It is well known that certain groups of individuals are at high risk of OBI. Patients on hemodialysis have a higher risk of parenterally transmitted infections.15 Due to their immunosuppressive state, the large number of transfusions they receive and the shared use of the dialysis machine with other patients, patients on hemodialysis are considered a risk group for OBI.15 The role of OBI in patients with chronic hepatitis C has been extensively studied.16 Coinfection with the hepatitis C virus (HCV) is associated with high OBI rates.16 In addition to these two risk groups, geographical areas considered endemic for HBV are correlated with the prevalence of OBI.17
The prevalence of this type of infection differs according to the pattern of endemicity of HBV infection in the region analyzed. The Brazilian Amazon is one of the regions of the world with the highest occurrences of hepatitis B, and HBV endemicity is not uniform; there are areas of high endemicity and areas with low and moderate endemicity for this virus.18-20 In this study, HBV DNA was detected in a sample from area with high endemicity for HBV in the state of Pará (Parauapebas municipality). Therefore, the expected prevalence of OBI was expected to be higher in populations with high exposure to HBV, i.e., those from areas with high endemicity.
The incidence of HBV/HCV coinfection has been reported and ranges from 1-15% worldwide.21 The prevalence of OBI varies among patients coinfected with HCV and is influenced by the geographic region, population and study design.21 Studies conducted in Spain, Italy, Japan, Taiwan, and Iran showed that approximately 10-15% of patients infected with HCV had HBV/HCV coinfection.22 In some Asian countries, more than 50% of patients had HBV/HCV coinfection.8 In the present study, HBV DNA was detected in six samples from individuals infected with HCV.
It is reported in the literature prevalence rates of OBI ranging from 0 to 58% in patients on hemodialysis.23 In Brazil, the prevalence of OBI among hemodialysis patients ranges from 0 to 15%.23 In the present study, HBV DNA was detected in two patients on hemodialysis, and the phylogenetic analysis identified genotype D in one of these patients.
A study involving 40 hemodialysis patients in the state of Santa Catarina found that genotype D (57.1%) was the most prevalent, followed by genotypes A (30.6%) and F (12.2%); in the state of Rio Grande do Sul, there was a higher prevalence of genotype D (60%), and genotypes A (34%) and F (5%) were also detected; in the state of Paraná, genotypes D (82.9%) and A (14.1%) were the most frequently identified.24 The literature suggests that genotype D is more closely linked to the spread of the virus in settings that provide hemodialysis. In addition, genotype D HBV isolates have been reported in outbreaks in hemodialysis units.25
Use of molecular tests with high sensitivity and specificity is recommended for diagnosing OBI, giving preference to the use of real-time PCR with a low limit of detection.6,10 Furthermore, it is also recommended that molecular analysis be performed in two or more different regions of the HBV genome.6,10 In this study, the partial regions of S, Pol and C were analyzed, but mutations associated with occult HBV infection were only identified in the S region.
In Brazil, HBV genotypes A, D, and F are the most prevalent, and their distributions differ by region. A multicenter study analyzed 1,004 samples from the North, Northeast, Central-West, Southeast and South geographic regions, covering 24 Brazilian states and seven HBV genotypes circulating in Brazil were identified.26 Genotype A was the most prevalent, identified in 589 (58.7%) of the samples, followed by genotypes D (23.4%), and F (11.3%).26
Genotype A was the most prevalent (71.6%), followed by genotypes D (14.2%) and F (10.9%) in Northern Brazil.26 In the present study, the phylogenetic analysis of isolates from patients with OBI identified genotypes A (A1) and D (D3 and D4). Although genotype D is not the most prevalent genotype in Brazil, it is present in all Brazilian regions. The D3 subgenotype originates from South Africa, Asia and Europe, and the D4 subgenotype originates from Oceania.4
Four mutations already described in the literature were identified in the S gene: E164D, I195M, P217L and P120S in this study. The preS/S gene are the most variable viral genome sequences. In particular, the “a” determinant domain, located between amino acids 99 and 170 of the S protein, is essential for the detection of HBsAg and the development of HBV vaccines. Several point mutations in this domain have been associated with occult HBV infection.3,27,28
A molecular study conducted in Rio de Janeiro identified 15 different S gene mutations associated with OBI.29 Of these, 12 substitutions (L104F, T116I, P120L, T126K, P127L, Q129H, M133T, F134R, P142I, K160R, Y161F, and V168A) were located in the “a” determinant. The other three mutations (L42P, L94S and P178Q) were identified outside this region.29
G145R mutation consists of a molecular change frequently found in the “a” determinant and is associated with occult HBV infection and the mutant virus's escape from the antibodies generated by the vaccine.27,28,30 In addition to the G145R mutation, positions 120, 126, 129, 130, 133, 134, 137, 140, 143, and 144 are also considered important points for the occurrence of` molecular changes associated with occult HBV infection.27,28,30
The substitutions E164D and I195M, identified in this study, are also considered resistance mutations to lamivudine, since these changes in HBsAg cause a change in the reading frame of the polymerase gene that results from the overlap between these two regions of the HBV genome.27,28,30 In the present study, all identified mutations associated with OBI were located in the S gene. Out of these mutations, two (E164D and P120S) were located in the “a” determinant, confirming the high mutation rate reported in the literature for this region of the HBV genome.
ConclusionsIn summary, the present study provides additional epidemiological and molecular information on Northern Brazil samples with a suggestive profile of occult HBV infection, in addition to reinforcing the importance of molecular diagnosis for this type of HBV infection, with a preference for the investigation of two or more regions of the viral genome and the use of molecular tests with high sensitivity and specificity. The molecular analysis also identified the circulating genotypes and mutations in the S gene, which are associated with the nondetection of HBsAg by commercial tests. New molecular studies with larger sample sizes that include other risk factors for OBI are essential to help establish the prevalence of this type of infection in Northern Brazil.
Authors’ contributionsPEBF: Conceptualization, Investigation, Writing original draft, Writing review & editing, SRA: Data curation, Formal analysis, Investigation, APM: Investigation, Methodology, VPS: Investigation, Methodology, HMN: Supervision, Funding acquisition.
The authors thank Edivaldo Costa Sousa Júnior from the Virology Department of the Evandro Chagas Institute for his assistance in the analysis of mutations and phylogeny. We thank American Journal Experts for the english translation of this manuscript. Finally, they thank researcher Dr. Manoel do Carmo Pereira Soares, former head of the Hepatology Department of the Evandro Chagas Institute, for his constant support in the development of studies in the field of hepatology. This research was financially supported by the Institutional Program for Scientific Initiation Scholarships of the Evandro Chagas Institute and the Hepatology/IEC Department in the performance of laboratory tests.