The purpose of this study was to identify the risk factors that predispose patients who are hospitalized with pressure ulcers (PUs) colonized by Gram-negative bacilli (GNB) to develop bacteremia. In addition, we also detected main phenotypes of resistance in infected and uninfected PUs. A prospective cohort study was conducted at the Clinical Hospital of the Federal University of Uberlândia including patients with Stage II or greater PUs, colonized or not with GNB, from August 2009 to July 2010. Infected ulcers were defined based on clinical signs and on positive evaluation of smears of wound material translated by a ratio of polymorphonuclear cells to epithelial cells ≥2:1, after Giemsa staining. A total of 60 patients with Stage II PUs were included. Of these 83.3% had PUs colonized and/or infected. The frequency of polymicrobial colonization was 74%. Enterobacteriaceae and GNB non-fermenting bacteria were the most frequent isolates of PUs with 44.0% of multiresistant isolates. Among patients who had infected PUs, six developed bacteremia by the same microorganism with a 100% mortality rate. In addition, PUs in hospitalized patients were major reservoir of multiresistant GNB, also a high-risk population for the development of bacteremia with high mortality rates.
Pressure ulcer (PUs) is a common clinical problem associated with high cost in terms of both treatment and human suffering.1 This is particularly common in elderly patients in general hospitals and home care settings, mainly in critical units where its incidence ranges from 8% to 40%.2
The microbiota of PUs is usually polymicrobial and complex and can be colonized with Gram-negative bacilli (GNB) multidrug-resistant and methicillin-resistant Staphylococcus aureus (MRSA). PUs can be reservoirs for resistant microorganisms3 and may evolve into local infections, also becoming a source of bacteremia in hospitalized patients.4
This study aimed to identify the risk factors that predispose hospitalized patients with PUs colonized by GNB to develop bacteremia by these microorganisms. In addition, we also detected the main phenotypes of bacterial resistance in infected and uninfected PUs.
Materials and methodsSetting and study designThe research was conducted at the Clinical Hospital of the Federal University of Uberlândia, a tertiary 510-bed teaching hospital, from August 2009 to July 2010. During the study period, 60 patients with Stage II or greater PUs were included in a prospective cohort study, whose medical records were identified by active surveillance and reviewed for demographic and risk factor data.
DefinitionsInfected PUThe presence of infection at the ulcer site was based on clinical signs and symptoms (i.e., erythema, edema, pain, foul odor, and purulent exudates, fever, delayed healing, discoloration of granulation tissue, friable granulation tissue, and wound breakdown).5,6 In addition, a PU was deemed infected when the smear of its material yielded a ratio of polymorphonuclear to squamous epithelial cells ≥2:1 after Giemsa staining.7
PU stagingPUs were classified according to the criteria proposed by Santos et al.8
BacteremiaDefined as the presence of viable bacteria in the blood documented by a positive blood culture.9 Only the first MRSA and GNB bacteremia episode was taken into consideration.
Multidrug-resistant (MDR) phenotypeThe criteria used for defining MDR phenotype was: non-susceptible to ≥1 agent of ≥3 antimicrobial classes.10
Specimen collection and microbiological techniquesPU cleaning was performed with sterile and warm saline solution (around 37.5°C) and an aseptic technique under irrigation pressure with a syringe (20ml) and needle (gauges 25X8-21).11 Afterwards, a sterile swab moistened with saline solution was rotated over a 1cm2 area of granulation tissue with sufficient pressure to press fluid out of the wound tissue.12
The clinical specimen was inoculated into MacConkey Agar, and the differentiation of Gram-negative microorganisms in Enterobacteriaceae family and non-fermenting bacilli was made through oxidation-fermentation (OF) and oxidase tests. For Enterobacteriaceae it was used glucose and lactose fermentation; indole production; motility; citrate; urea hydrolysis; sulfidric gas production; phenylalanine deaminase; lysine and ornithine decarboxylase; methyl red reaction; and Voges-Proskauer test. For non-fermenting GNB it was used nitrate reduction; gluconate use; pigment production; lysine decarboxylase activity; urease activity; indole production; acetamide and esculin hydrolysis. The swab was inoculated into Mannitol Agar, and Staphylococcus aureus was identified as coagulase-positive by classic techniques.13
Selected S. aureus and GNB isolates were evaluated using the disk diffusion test for resistance to methicillin and fluoroquinolones, and to third and fourth generation cephalosporins and carbapenems, respectively. Quality-control protocols were used according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2014).14
Ethical aspectsEthical approval was obtained from the Ethics Committee of Uberlandia Federal University according to the requirements of the Ministry of Health, under protocol numbers 333/09 and 118/05.
Statistical analysisTo assess the association of risk factors with bacteremia, patients with infected and uninfected PU were compared using the Student's t-test for continuous data and the Fisher exact test or chi-square test for categorical data, when appropriate. A p-value less than or equal to 0.05 was considered significant. Statistical analyses were performed with GraphPad Prism version 4 (GraphPad Software, San Diego, CA).
ResultsA total of 60 patients were investigated. 117 patients were excluded due to Stage I PUs or because the patients and/or family members have not agreed to participate in this study. The patients were predominantly men (70.0%), mean age of 61 years with a wide range (20–88 years). The majority of patients were hospitalized for clinical reasons (53.3%) or trauma (26.7%). Only six (10.0%) patients were admitted because of infected PUs.
The patients were hospitalized for an average of 103 days (SD±84.8 days). Cardiomyopathy (78.3%) and diabetes mellitus (43.3%) were the most common comorbidities. Gastrointestinal catheter (85.0%), central venous catheter (55.0%), mechanical ventilation (45.0%), urinary catheter (40.0%), three or more invasive devices (55.0%), and three or more classes of antimicrobial agents (77.9%) were used by most of the patients. The hospital mortality rate was also high (41.7%) in this cohort (data not shown).
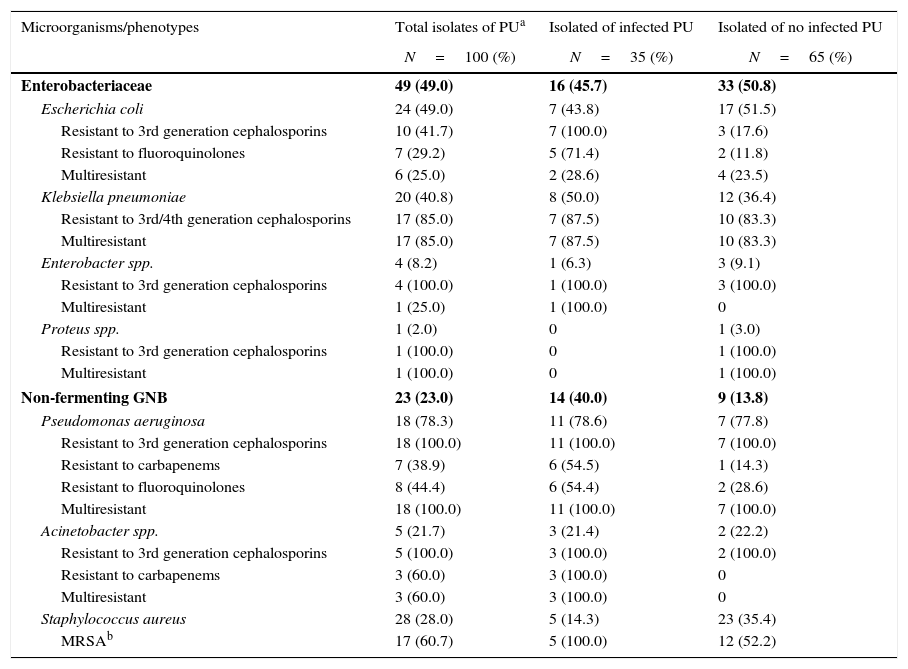
Enterobacteriaceae were the most commonly isolated (49.0%) group of bacteria in colonized/infected patients with PUs, followed by Staphylococcus aureus (28.0%) and non-fermenting GNB (23.0%), mostly Pseudomonas aeruginosa and Acinetobacter spp. Colonization with multiresistant bacteria including Klebsiella pneumoniae (17/20, 85.0%), Escherichia coli (6/24, 25.0%), Enterobacter (1/4, 25.0%), Pseudomonas aeruginosa (18/18, 100.0%), Acinetobacter spp. (3/5, 60.0%), and methicillin-resistant Staphylococcus aureus (17/28, 60.7%) were the predominant bacteria in our patients. The frequency of epidemiologically relevant phenotypes such as fluoroquinolones- and carbapenems-resistant Pseudomonas aeruginosa, carbapenems-resistant Acinetobacter spp, third generation cephalosporins-resistant Klebsiella pneumoniae, and Escherichia coli were surpassed 40% among GNB isolates. Among the 16 patients with infected PUs, 35 samples were isolated of which 29 (82.9%) were multiresistant bacteria, whereas among 34 patients with non-infected PUs, 65 samples were isolated, of which 35 (52.3%) were multiresistant (Table 1). By univariate analysis, no risk factors were statistically associated with GNB bacteremia in patients with Stage II or greater PUs colonized by these pathogens (data not shown).
Microorganisms and resistance phenotypes in isolated pressure ulcers.
| Microorganisms/phenotypes | Total isolates of PUa | Isolated of infected PU | Isolated of no infected PU |
|---|---|---|---|
| N=100 (%) | N=35 (%) | N=65 (%) | |
| Enterobacteriaceae | 49 (49.0) | 16 (45.7) | 33 (50.8) |
| Escherichia coli | 24 (49.0) | 7 (43.8) | 17 (51.5) |
| Resistant to 3rd generation cephalosporins | 10 (41.7) | 7 (100.0) | 3 (17.6) |
| Resistant to fluoroquinolones | 7 (29.2) | 5 (71.4) | 2 (11.8) |
| Multiresistant | 6 (25.0) | 2 (28.6) | 4 (23.5) |
| Klebsiella pneumoniae | 20 (40.8) | 8 (50.0) | 12 (36.4) |
| Resistant to 3rd/4th generation cephalosporins | 17 (85.0) | 7 (87.5) | 10 (83.3) |
| Multiresistant | 17 (85.0) | 7 (87.5) | 10 (83.3) |
| Enterobacter spp. | 4 (8.2) | 1 (6.3) | 3 (9.1) |
| Resistant to 3rd generation cephalosporins | 4 (100.0) | 1 (100.0) | 3 (100.0) |
| Multiresistant | 1 (25.0) | 1 (100.0) | 0 |
| Proteus spp. | 1 (2.0) | 0 | 1 (3.0) |
| Resistant to 3rd generation cephalosporins | 1 (100.0) | 0 | 1 (100.0) |
| Multiresistant | 1 (100.0) | 0 | 1 (100.0) |
| Non-fermenting GNB | 23 (23.0) | 14 (40.0) | 9 (13.8) |
| Pseudomonas aeruginosa | 18 (78.3) | 11 (78.6) | 7 (77.8) |
| Resistant to 3rd generation cephalosporins | 18 (100.0) | 11 (100.0) | 7 (100.0) |
| Resistant to carbapenems | 7 (38.9) | 6 (54.5) | 1 (14.3) |
| Resistant to fluoroquinolones | 8 (44.4) | 6 (54.4) | 2 (28.6) |
| Multiresistant | 18 (100.0) | 11 (100.0) | 7 (100.0) |
| Acinetobacter spp. | 5 (21.7) | 3 (21.4) | 2 (22.2) |
| Resistant to 3rd generation cephalosporins | 5 (100.0) | 3 (100.0) | 2 (100.0) |
| Resistant to carbapenems | 3 (60.0) | 3 (100.0) | 0 |
| Multiresistant | 3 (60.0) | 3 (100.0) | 0 |
| Staphylococcus aureus | 28 (28.0) | 5 (14.3) | 23 (35.4) |
| MRSAb | 17 (60.7) | 5 (100.0) | 12 (52.2) |
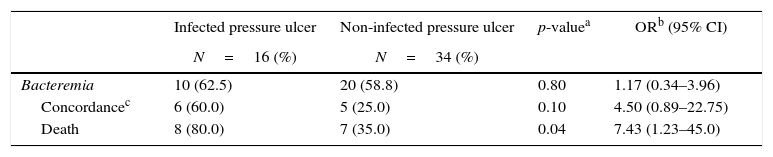
Of the 50 patients (83.3%) with PU colonized by GNB, 16 (32.0%) developed local infections. Out of these 16 patients with infected ulcer, 10 patients (62.5%) developed bacteremia. Among these 10 patients, six had the same isolates recovered from both exudate and blood with similar phenotype/antibiotic susceptibility pattern. The mortality in this group of patients with PU infected with GNB bacteremia was higher (OR: 7.43, 95% CI, 1.23–45.0, p=0.04) than in patients with non-infected PUs (Table 2).
Bacteremia in patients with pressure ulcers colonized by Gram-negative bacilli.
Pressure ulcers are a public health problem that results in distress and disability, and constitute a great challenge to health administrators. They are particularly common among elderly patients in general hospitals and home care settings.2
Our study included 60 patients with Stage II or greater PUs with characteristics of critically ill patients, the majority with three or more invasive procedures (55.0%) and two or more comorbidities (75.0%). In addition, these patients had a rather long length of hospital stay (mean 103 days) and a high hospital mortality rate (41.7%). Stage III PUs were the most common (32.0%) in our series.
PUs are also frequently colonized by several species of bacteria and surface cultures yield a polymicrobial flora of both Gram-positive and Gram-negative aerobic and anaerobic species.15 The microorganisms most commonly isolated in chronic wounds are usually Staphylococcus aureus, β-hemolytic Streptococci, Enterococcus spp., aerobic GNB Enterobacteriaceae and Pseudomonas spp., and in particular, resistant bacteria such as methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus spp., ciprofloxacin-resistant Pseudomonas aeruginosa, and extended-spectrum beta-lactamase (Escherichia coli and Citrobacter).16,17 In our study, 50 out of 60 patients (83.3%) had PUs with cultures positive for GNB, most with mixed flora (74.0%). Most isolates were identified as Enterobacteriaceae (49.0%), Escherichia coli (49.0%) and Klebsiella pneumoniae (40.8%), and non-fermenting GNB (23.0%), mainly Pseudomonas aeruginosa (78.3%), and Staphylococcus aureus (28.0%). A total of 63% of the isolates were multiresistant to different antibiotics, including Pseudomonas aeruginosa (100.0%), Proteus spp. (100.0%), Klebsiella spp (85.0%), MRSA (60.7%), Acinetobacter spp (60.0%), Escherichia coli (25.0%), and Enterobacter spp. (25.0%).
One of the few reports in the literature about the role of polymicrobial synergy in local wound infection pointed out that the recovered bacteria from PUs are similar to those of acute necrotizing soft tissue infections, namely anaerobic and aerobic bacteria, thus likely contributing to the deterioration of a lesion.18 Our results confirming that the polymicrobial flora was observed in the majority (74.0%) of infected wounds are in line with the findings of that report.
The association of PUs with bacteremia is well established, but few studies have addressed this problem systematically. PUs may serve as occult foci for blood stream infection, the most common major complication of PUs. Infected PUs are more likely to cause bacteremia than wounds only colonized with polymicrobial microorganisms,17 also evident in our results.
In hospitalized patients, the relationship between bacteremia and PU has been associated with 50% mortality rate.19 Among 21 patients with sepsis attributed solely to PU, bacteremia was documented in 16 (76%) cases, and 47.6% of the patients died, eight of these despite appropriate antibiotics treatment.20 Our data showed that out of 16 patients with infected PUs, 10 (62.5%) developed GNB bacteremia, six had the same microorganism isolated from the PU and blood. Out of these six, Pseudomonas aeruginosa was recovered from four patients and Klebsiella pneumoniae from two.
In our study, the 16 patients with bacteremia caused by the same GNB that was recovered from the PU had a higher risk of dying (OR=7.43; p≤0.04) when compared with 34 patients who did not have infected PUs and were just colonized by these microorganisms. The hospital mortality rate was 80.0% in patients with infected PUs who developed bacteremia, compared to 35.0% in those who developed bacteremia, but had no infected PUs in spite of the presence of the same microorganism in PUs and blood.
In conclusion, Stage II or greater PUs in hospitalized patients are reservoirs of multiresistant GNB, such as Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Colonization of PUs was predominantly polymicrobial with significant association with infection in other anatomic sites. This study also emphasizes the importance of PUs as a potential source of bacteremia in hospitalized patients leading to high mortality rates.
Conflicts of interestThe authors declare no conflicts of interest.
We thank the patients of Clinical Hospital of Federal University of Uberlandia who consented to participate in the study and the staff members for their cooperation. We also thank to Deivid W.F. Batistão for helping in statistical analysis and the Brazilian Agency FAPEMIG and CNPq for financial support.