Despite vaccine development and vaccination programs underway around the globe, the coronavirus disease 2019 (COVID-19) pandemic has not been controlled as the SARS-CoV-2 virus is evolving and new variants are emerging. This study was conducted to sequence and molecularly characterize the representing samples from the early fourth SARS-CoV-2 wave in Iraq. Here, we have performed next-generation sequencing of whole-genome sequencing of two representing samples from the country's early beginning of the fourth pandemic wave. The samples were sequenced using Illumina Miseq system, and the reference sequences were retrieved from GISAID database. Phylogenetic analysis was performed through Mega software. This study provides an initial sequence analysis and molecular characterization of the first Omicron variant cases recorded in the country. Our analysis revealed many mutations on the spike glycoprotein, especially on the receptor binding domain, with potential impact on immune escape and infectivity. The study findings suggest considering the highly mutated immunogenic epitope of the Omicron variant as a reference for developing a new vaccine for combating the ongoing pandemic.
Approximately two years have passed since Wuhan's first contagious agent was reported. The pandemic coronavirus disease 2019 (COVID-19), caused by the positive sense single-stranded RNA virus denoted as SARS-CoV-2.1 Despite the continuous emergence of new variants of the virus and the short-lived nature of vaccine-induced immunity, the peak of the daily new cases was declining, and the results raised hope that the coronavirus pandemic could be contained. Consequently, the current pandemic could get one step closer to an end after vaccine development.2 Though globally, COVID-19 led to significant burden of illness and unpredictable death in different parts of the world.1
Presently, the world faces another recent variant that led researchers to attempt to collect primary data on that new variant initiated pathogenesis and its mechanisms. It has been revealed that Omicron variants with new mutations could affect the transmissibility of the virus and the host's immunity.3
The SARS-CoV-2 Omicron variant was first recorded in South Africa on November 24th, 2021, and was assigned as a variant of concern (VOC) within two days by the World Health Organization (WHO).4 The Omicron variant has the largest number of mutations among all other SARS-CoV-2 variants, harbouring more than 50 mutations on the virus genome. S gene was found to be the most mutated gene, and 15 of those mutations are on the receptor-binding domain (RBD) of the virus.5 The S gene codes for Spike glycoprotein that is used by the virus to interact with the cellular angiotensin converting-2 enzyme (ACE2).6 It was suggested that such mutations in the Spike glycoprotein might lead to escape from the developed immune response due to infection or vaccination and may cause many breakthrough infections or reinfection with mutated viral strains.7,8
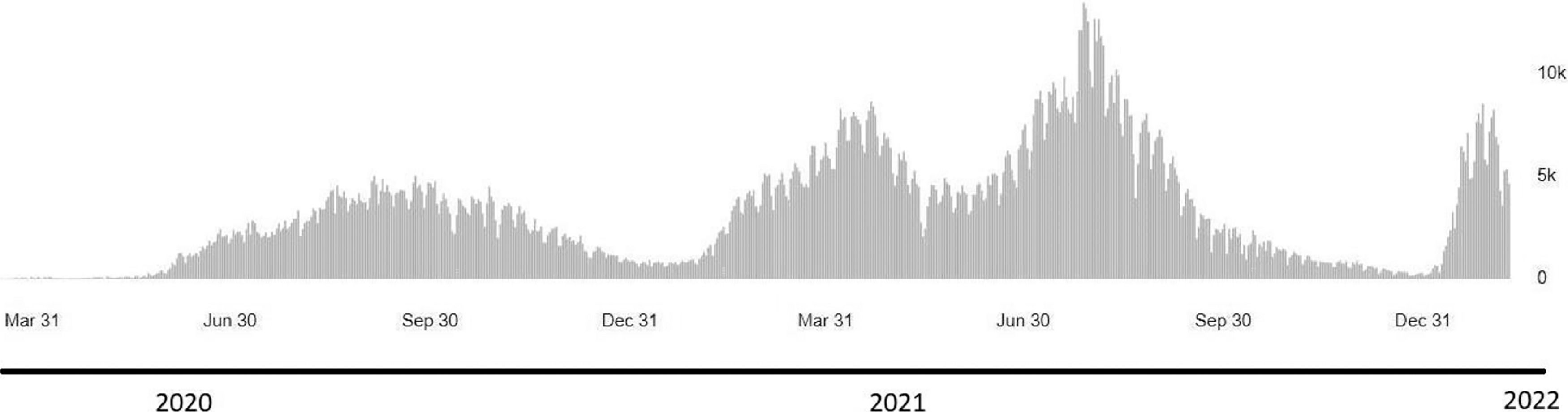
Like other places, as of January 25th, 2022, the Kurdistan region had accumulated laboratory-confirmed cases of SARS-CoV-2 omicron variant; presently, it is considered the fourth wave of infection. The first wave started in early March 2020 and was followed by a wave that topped in July 2020 and ended in September.9 The second wave peaked in January 2021 and ended in March 2020. The third wave, which peaked in July and ended in September 2021, was overwhelmed by the Delta variant. In late December 2021, the Omicron variant was detected in Duhok Province and associated with rapidly increasing cases; the Kurdistan region is in the red zone of infection.
However, the number of shared sequences diagnosed from Iraq in the international database is still scarce (GISAID, https://www.gisaid.org/). Mutations of SARS-CoV-2 may happen over time to escape the host immune response, especially with the robust immune pressure in humans.10 This possibly leads to the emergence of new virus variants with higher infectivity, transmissibility, and virulence potentials.11
Herein, we aimed to perform whole-genome sequencing for the samples of SARS-CoV-2 from the early begening of the fourth wave of the pandemic in Iraq, in order to characterize the mutation patterns of the Omicron variant in the country.
Materials and methodsSamplesAfter the emergence of the omicron variant in South Africa and its rapid spread to other countries worldwide, the daily cases in the Kurdistan region started to surge again; this has brought about the need to sequence representative new cases and identify the circulating variant and its mutation patterns. Two positive samples that were collected on December 21st 2021for routine SARS-CoV-2 testing at Duhok Central public health laboratory have been selected for whole-genome sequencing.
Viral RNA extraction and Real-Time PCRThe selected samples were subjected to extraction and quantitative detection using RNA extraction and real-time PCR-based virus detection system QIAprep & amp Viral RNA UM Kit (Qiagen). Ct value of selected samples was (< 20) as demanded by the sequencing laboratory. The RNA was then extracted from the viral transport medium by QIAamp Viral RNA Mini Kit (Qiagen), and transported to the Intergen commercial company (Ankara, Turkey) on dry ice for whole-genome sequencing.
SARS-CoV-2 Next Generation Sequencing (NGS) Bioinformatics AnalysisThe samples were retested for target confirmation and RNA integrity following arrival at Intergen. Ipsogen RT Kit and nanomere primers from (Qiagen, Germany) were used for reverse transcription and cDNA synthesis. Indexing and tagmentation of the individual samples was performed using illumina sample preparation kit. Next-generation sequencing was performed using illumina Miseq instrument and equipment (Illumina, San Diego, CA, USA) following manufacturers' instructions. IGV 2.8.9 (Broad Institute) software was used to evaluate sequencing data. The short reads sequencing was assembled and aligned with the reference genome (NC_045512.2) through BWA-MEM alignment algorithm.12 The annotation of the assembled consensus sequences was completed by Annovar.13 Together, Lofreq (version 2) was used for the identification of mutation and variant calling.14 The accepted sequencing read underwent quality control. The sequences had coverage of (> 99%) and gap length of (< 30) bps that were subjected to further analysis. The obtained whole-genome sequence was submitted to the GISAID database, and the assigned accession numbers were received (EPI_ISL_9084009, EPI_ISL_9765388).
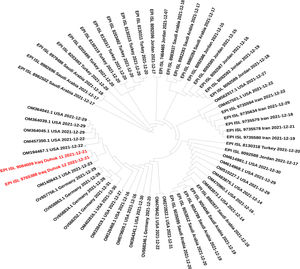
Variants, lineage and phylogenetic analysisThe whole-genome sequences of this study (n = 2), samples from the surrounding countries, and blast search result sequences around the world have been retrieved from GISAID and the National Centre for Biotechnology Information data banks. The criteria considered for sequence selection were good quality sequences, without running Ns, collected during December 2021, and the characterized as Omicron variants. The developed dataset has been subjected to quality control, and low-quality sequences have been excluded. The phylogenetic tree was constructed to reveal the relationship between the sequences of this data set and track down the source of the emerging Omicron variant in Iraq. The phylogenetic tree was built using the neighbour-joining method implemented in the Molecular Evolutionary Genetics Analysis program (MEGA 11).15 The variants and linage identification were made using the Nextclade sequence analysis and Pangolin system version (v3.1.14), respectively.16,15
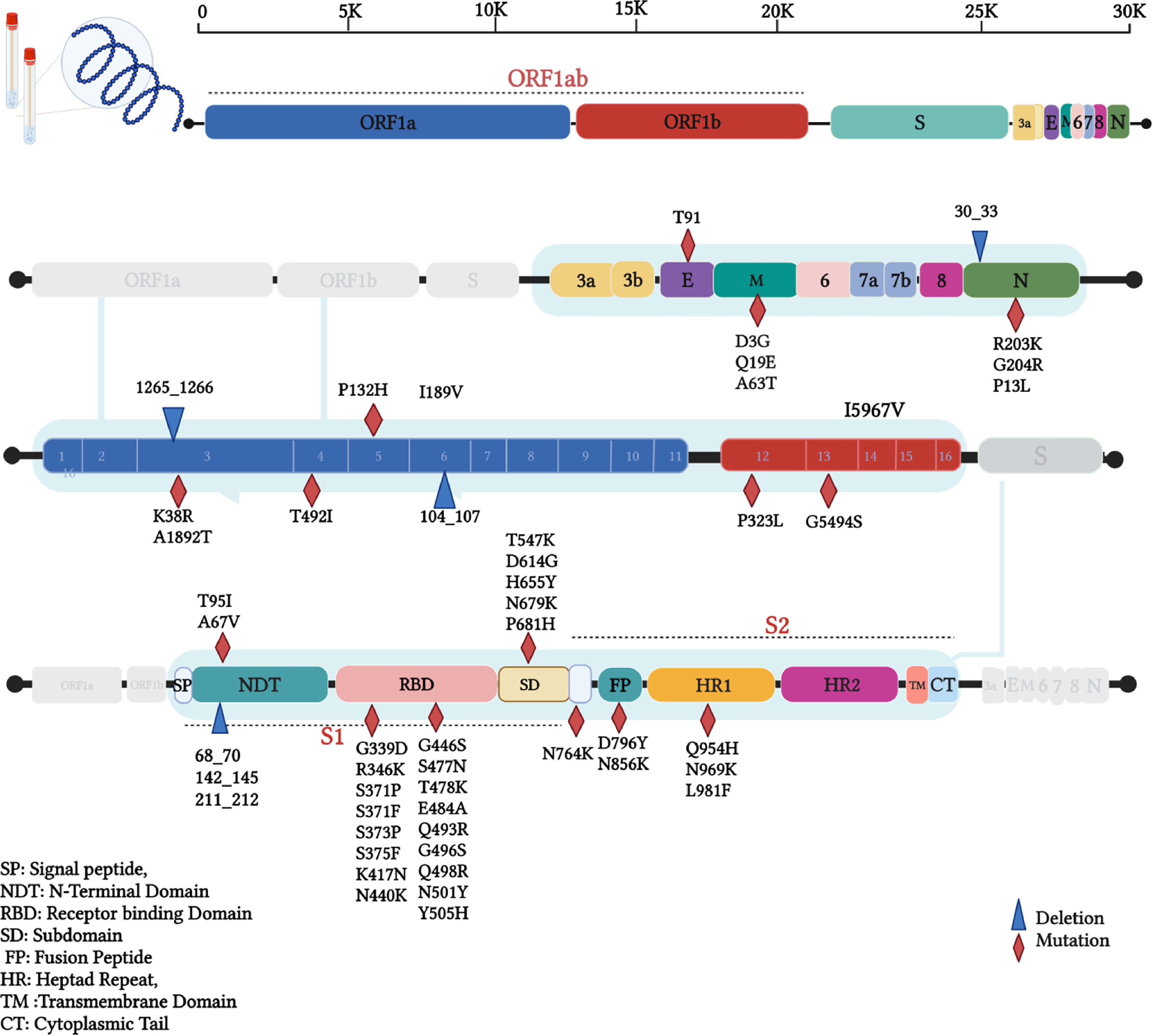
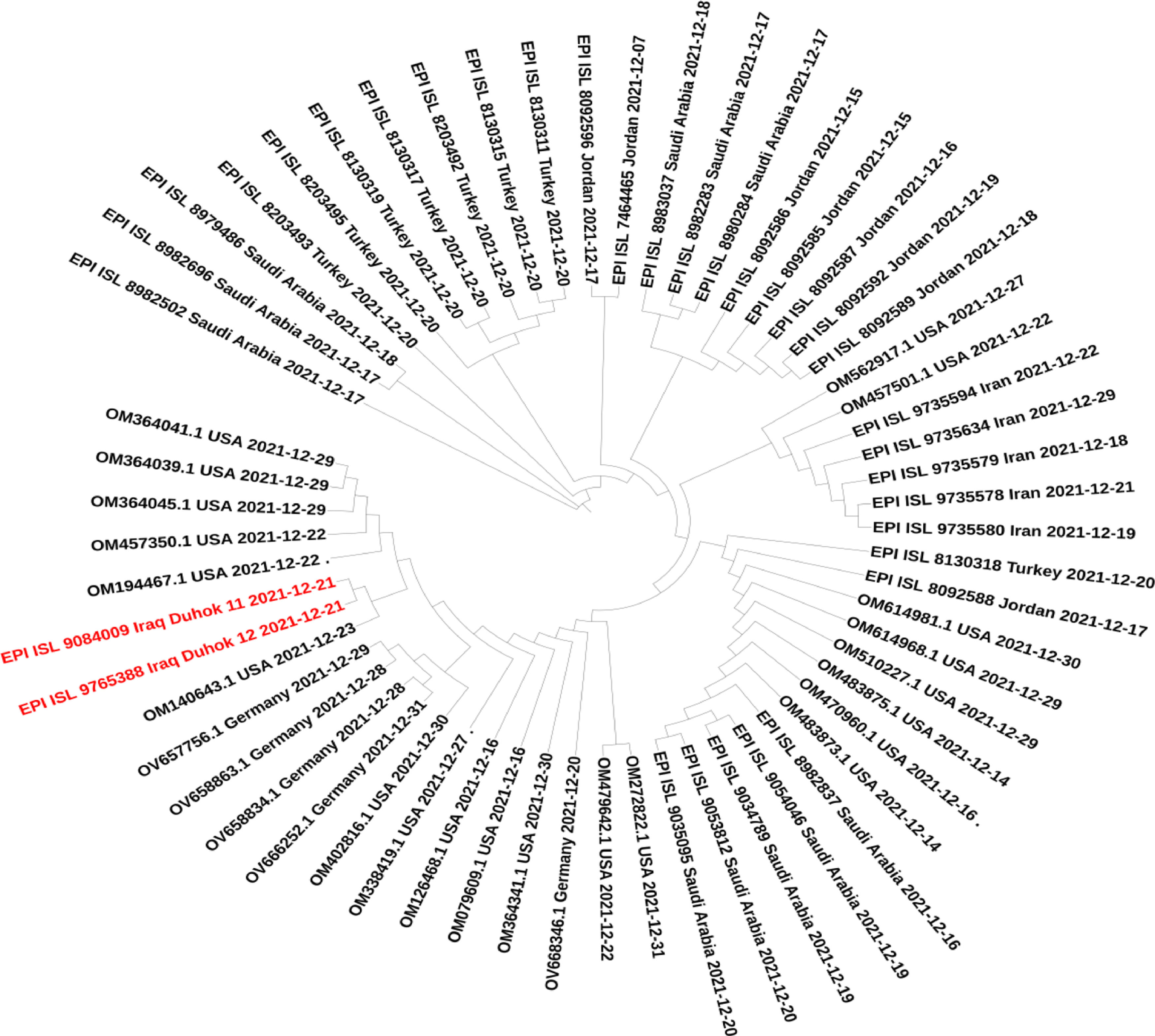
ResultsTwo isolates of this study were found to be Omicron VOC using Nextclade platform,16 and the Pangolin tool has identified these sequences as BA.1.1 lineage of Omicron.15 The mutation analysis has revealed that Iraqi isolates of this study have a total of 62 different mutations; among these, a total of 45 mutations were non-synonymous, 10 synonymous, one upstream, and deletions at six different positions that resulted in the deletion of 39 single nucleotides (Table 1). Interestingly, 34 (54.8%) of these mutations were found on Spike protein, 33 non-synonymous mutations, followed by 15 (24.2%) mutations in the ORF1ab region that constituting of total mutations. Other genomic regions N, ORF3a, E, M, ORF6, and ORF7b were less frequently mutated. Non-synonymous mutations on the SARS-CoV-2 genome revealed that the mutations hot spot was Spike protein (Fig. 1). The synonymous mutations at nucleotide position 3373 C > T and 16744 G > A were found to be a signature of the two isolates of our study that were not found in the tested data set. The phylogenetic analysis for the two isolates of this study and retrieved sequences from both GISAID and NCBI databases revealed several sub-cladding sequences of the tested data set (Fig. 2). The sequences of this study were found to group with isolates from USA making one to speculate that active international travel could have been the potential source of the Omicron found in Iraq.
Total mutations, effect on amino acid, and distribution on genomic positions of SARS-CoV-2 (Omicron variant) isolates of this study.
In a time of global efforts to develop antivirals and distribute the vaccine, daily cases are rising due to the emergence of new variants in some countries. While the delta variant remains circulating worldwide, the recently discovered Omicron VOC has revealed a tendency to become the upcoming dominant variant. In this study, we are showing a sequence analysis and the mutation pattern of the first Omicron variant sequences from cases recorded in Iraq in December 2021 that subsequently led to the announcement of the fourth SARS-CoV-2 wave in the country by the Ministry of Health (Fig. 3).
The analyzed Omicron VOC of SARS-CoV-2 sequences of this study showed many mutations on S-gene, followed by ORF1ab, N-gene, M-gene. ORF3a, E-gene, ORF6, and ORF7b were the lowest mutated genes. The most mutated S-gene, which encodes a structural protein, functions as a viral binding protein to the host cell receptors and determines the host range.17 A total of 30 non-synonymous mutations have been found on this gene, including A67V, T95I, G339D, R346K, S371P, S371F, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F, and only D1146D as synonymous mutation along with 68_70del, 142_145del, and 211_212del deletions. This gene was reported to have 4-5 times more mutation potential compared to other genomic positions.18
It remains uncertain how the Omicron VOC has acquired such vast number of mutations, specifically on the Spike protein. Since this variant was first detected in an immunocompromised patient in South Africa, less selective pressure was on the virus. Still, the prolonged duration of the infection could be the reason for such a huge evolution. In general, an increasing and persistent selective pressure on the virus on both population and individual levels speed up the viral evolution process. The dominant variant D614G in the Spike gene has also been conserved for the Omicron variant; this variant has been found in all other VOCs,19 which is related to increased transmission and infection rates and viral escape from reactive antibodies.20 The receptor-binding domain (RBD) of the spike protein, which is a primary target for neutralizing antibodies, has been shown to be a mutation hot spot subunit within the Spike glycoprotein having 17 mutations, including G339D, R346K, S371P, S371F, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H. Novel mutations of RBD of Omicron were G339D, R346K, S371P, S371F, S373P, S375F, N440K, G446S, Q493R, G496S, Q498R, Y505H mutations, in addition to other mutations that had been acquired from previous VOCs. Mutations like K417N, N440K, G446S, T478K, E484A, and H69/V70 deletions found previously in other VOCs have been associated with escape from the neutralizing antibodies and immune escapes.5 Furthermore, Omicron has many novel mutations all over the spike protein like N-terminal domain (NTD), furin cleavage site, and S2 subunit that could potentially affect binding to ACE2 receptor and reaction to antibodies based on results from previous reports.21,22
ORF1ab, which constitutes 2/3 of the entire genome, was reported to be a mutation hot spot in all other VOCs,23,24 while the Omicron sequences of this study in this region were found to be less mutated (24.2%) compared to Spike protein. The ORF1ab gene functions in viral replication by coding for a helicase protein and RNA-dependent RNA polymerase enzyme.25 Among the non-structural proteins of ORF1ab, nsp3 was most mutated with K38R and A1892T variants and deletion at 1265_1266del amino acid positions. Variant T492I was detected on nsp4 and I189V with non-frame shift deletion (104_107del) on nsp6. These mutations and deletions could affect replication and evolution processes. It is believed that nsp3, nsp4, and nsp6 are responsible for double-membrane activation that enables transcription and replication activity26 and allows the evolution process due to selective pressure on the virus.27
N gene encodes the nucleocapsid protein, which is also involved in the replication and transcription process.28 This protein has conserved mutations in three consecutive nucleotides at position 28881-28883 (GGG > AAC) that resulted in a double amino acid change (R203K G204R); these variants were detected at early waves of the pandemic and have been conserved in subsequent emerging variants around the world.23,29,30 Nevertheless, ORF6 and ORF7 are accessory proteins that have a role in suppressing innate immunity, signalling pathways, and interferon (IFN) expression.31 Duhok sequences have shown less frequent mutations on these two proteins, and the two nucleotide changes (27259 A > C and 27807 C > T) were silent; these data are in line with previously analyzed sequences of the other VOCs.32
Interestingly, a silent mutation 3373 C>T was found to be a signature of our two sequences and had not been identified in any other sequences of our data set. At the same time, non-synonymous mutation G5494S of our isolates was found as a notable change only found in sequences from USA and Germany. Nevertheless, phylogenetic analysis of our data set has shown that our isolates were clustered with Omicron sequences from the USA, suggesting the source of the first Omicron emergence in the country through active international travelling.
This high number of mutations on the SARS-CoV-2 genome, specifically on spike protein, could increase viral transmission and immune escape compared to other VOCs. Furthermore, the accumulation of these bulky mutations on the immunogenic epitopes of Spike protein brought about the necessity of developing new vaccines that include the Omicron variant as a potential reference strain. Meanwhile, further studies are required to identify the infectivity and effectiveness of available vaccines against the Omicron variant.
In conclusion, this study provides an analysis of the first recorded Omicron variant sequences in Iraq that have caused a sharp increase in daily cases and the fourth wave of the pandemic announcement in the country. This work represents the first publication of Omicron sequence analysis in the country. One mutation was a unique signature of our isolates, in addition to a rare mutation only found in two sequences from the USA and Germany. These findings provide data on the mutation pattern in circulating variants in the country, which could help public health authorities in issuing and upadating the control roles for SARS-CoV-2 emergence. Here we recommend future studies focus on the impact and function of such genomic variants on the virus's infectivity, pathogenesis, and severity. Nevertheless, developing a new vaccine, especially multivalent vaccines containing multiple VOCs, could be a good step forward in controlling the latest infections.
Ethics approvalEthical Committee of the Directorate General of Health in Duhok has approved this study. The next-generation sequencing of the SARS-CoV-2 positive samples has been done after the cases underwent regular SARS-CoV-2 diagnostic tests at Duhok Central public health laboratory.
Availability of data and materialsThe SARS-CoV-2 sequences of this study are deposited in the Global Initiative on Sharing all Individual Data (GISAID).
FundingNo funding has been received.
The authors acknowledge the Duhok Central public health laboratory for the valuable support of this study.