To investigate the relationship between the polymorphism of human leukocyte antigen (HLA)-DRB1 and the susceptibility and repellency of drug use combined with HIV infection in Chinese.
MethodsA total of 213 unrelated healthy people, 41 HIV-infected drug users, 24 HIV-uninfected drug users, and 64 HIV-infected non-drug users were recruited. Their HLA-DRB1 allele frequencies were analyzed by PCR-SSP and allele distribution was analyzed.
ResultsCompared with healthy controls, in drug users, the frequencies of HLA-DRB1 *0401-041, *1001 were significantly higher; in HIV-infected patients, the frequencies of HLA-DRB1 *0101-0103, *0401-0411, *1001 were significantly higher, while the frequencies of DRB1 *1501-1502, *1101-1105, *1301-1302, DRB4, DRB5 were significantly lower; in HIV-infected drug users, the frequencies of HLA-DRB1 *0101-0103, *0401-0411, *0801-0806, *1001, *1401/1404/1405 were significantly higher, while the frequencies of DRB1 *1301/1302, 1501-1502, DRB5 were significantly lower.
ConclusionThere is close relationship between the polymorphism of HLA-DRB1 alleles and drug use with HIV infection, which plays an important role in elucidating the pathogenesis and providing the basis for therapeutics and prophylaxis of patients with drug use and HIV infection.
Drug abuse is a serious social problem. By October 2011, recorded drug users in China has reached 1.7 million, of which the number of the death due directly to drug abuse is 50,000. Actual death toll may be even higher. Many complications accompany with drug users, of which HIV infection is the most common. Individuals show significant difference in drug use combined with HIV infection, which mainly depends on human major histocompatibility complex (MHC). Human leukocyte antigen (HLA), the gene product of MHC, is a genetic system which is first discovered related to disease.1–3 As an important gene cluster related to immune system function in humans, different genotypes of HLA complex may influence the susceptivity and repellency of drug use combined with HIV infection. HLA complex, a group of closely linked gene cluster, exists in short arm of the human sixth chromosome. The codogenic HLA antigen determines the reject reaction of organism, and is related to immune response and immunological regulation. HLA complex is divided into HLA-I, HLA-II and HLA-III according to different gene product and gene function, and is coded as antigen HLA-I, antigen HLA-II and antigen HLA-II. Class II gene is named as HLA-D, which can classified into HLA-DR, HLA-DQ, HL-DP and other subregions, mainly expressed in immunocell. They can be functioned as labeled molecules among immunocells to activate immune response and regulate the interaction of immunocells, and plays an important role in immune response.4–7 For the high polymorphism of HLA complex, same phenotype in every individual is very rare, thus HLA becomes an important target in the investigation of many diseases, including autoimmune disease, infectious diseases, tumor and so on.8–12 High expression of HLA-DR is a symbol of T cell activation. According to statistics made by International Histocompatibility Working Group (IHWG), the number of alleles among noetic DRB1 sites in antigens HLA-II was 494, and this polymorphism was the main genetic factor13,14 to induce immune response in different individuals and different susceptivity to disease among the group. In the study, polymerase chain reaction-sequence specific primers (PCR-SSP) was used to test HLA-DRB1 genotype among 41 HIV-infected drug users, 24 HIV-uninfected drug users, and 64 HIV-infected non-drug users, and compared with HLA-DRB1 genotype of 213 healthy people, to explore the relationship between gene polymorphism of HLA-DRB1 and drug use combined with HIV infection.
In the pilot study, 41 HIV-positive drug users (average age 37±16), including 27 males and 14 females recruited from Centre for Disease Control and Prevention (Hubei, China) and Dali Drug Rehabilitation Center (Yunnan, China), were confirmed as HIV infectors by Western blot. 24 HIV-uninfected drug users (average age 30±11), including 12 males and 12 females, were recruited from Dali Drug Rehabilitation Center (Yunnan, China). 64 HIV-positive non-drug users (average age 42±9), including 44 males and 20 females, were recruited from Wuhan, Qichun, Songzi, Huangshi and nearly ten countries and cities by Centre for Disease Control and Prevention (Hubei, China), and were confirmed as HIV infectors by Western blot. 213 healthy people (average age 39±11), including 138 males and 75 females, were healthy people or healthy blood donors who had no blood relationship and were selected randomly from Zhongnan Hospital of Wuhan University, Wuhan Central Blood Bank, Wuhan Third Hospital, and Tongji Hospital of Huazhong University of Science and Technology. 5mL of anticoagulated blood was collected respectively, and stored at 4°C.
HLA-DRB1 allele sequence specific primers were designed based on the reference sequences from GenBank database, and were synthesized by Shanghai Sangon Biological Engineering Technology And Services Co., Ltd. (Tables 1 and 2).
HLA-DRB1 allele-specific primers.
| 5′-Primers | Sequence5′→3′ | 3′-Primers | Sequence5′→3′ | Size of PCR product (bp) | DRB primer amplified specificity |
|---|---|---|---|---|---|
| 5′01 | TTGTGGCAGCTTAAGTTTGAAT | 3′047 | CTGCACTGTGAAGCTCTCAC | 255 | 0101-0102 |
| 3′048 | CTGCACTGTTGAAGCTCTCCA | 255 | |||
| 5′01 | TTGTGGCAGCTTAAGTTTGAAT | 3′10 | CCCGCTCGTCTTCCAGGAT | 130 | 0103 |
| 5′02 | TCCTGTGGCAGCCTAAGAG | 3′01 | CCGCGCCTGCTCCAGGAT | 197 | 1501-1502 |
| 5′02 | TCCTGTGGCAGCCTAAGAG | 3′02 | AGGTGTCCACCGCGGCG | 213 | 1601-1602 |
| 5′03 | TACTTCCATAACCAGGAGGAGA | 3′03 | TGCAGTAGTTGTCCACCCG | 151 | 0301-0302 |
| 5′06 | GACGGAGCGGGTGCGGTA | 3′048 | CTGCACTGTGAAGCTCTCCA | 217 | 0301 |
| 5′03 | TACTTCCATAACCAGGAGGAGA | 3′047 | CTGCACTGTGAAGCTCTCAC | 189 | 0302, 1302, 1305, |
| 1402, 1403, 1409 | |||||
| 5′04 | GTTTCTTGGAGCAGGTTAAACA | 3′048 | CTGCACTGTGAAGCTCTCCA | 260 | 0401-0411 |
| 3′047 | CTGCACTGTGAAGCTCTCAC | 260 | |||
| 5′07 | CCTGTGGCAGGGTAAGTATA | 3′079 | CCCGTAGTTGTGTCTGCACAC | 232 | 0701-0702 |
| 5′08 | ACTACTCTACGGGTGAGTGTT | 3′05 | CTGCAGTAGGTCTCCACCAG | 214 | 0801-0806 |
| 5′09 | GTTTCTTGAAGCAGGATAAGTTT | 3′079 | CCCGTAGTTGTGTCTGCACAC | 236 | 0901 |
| 5′10 | CGGTTGCTGGAAAGACGCG | 3′047 | CTGCACTGTGAAGCTCTCAC | 204 | 1001 |
| 5′05 | GTTTCTTGGAGTACTCTACGTC | 3′06 | CTGGCTGTTCCAGTACTCCT | 176 | 1101-1105 |
| 5′08 | ACTACTCTACGGGTGAGTGTT | 3′08 | CACTGTGAAGCTCTCCACAG | 248 | 1201-12-2 |
| 5′03 | TACTTCCATAACCAGGAGGAGA | 3′10 | CCCGCTCGTCTTCCAGGAT | 130 | 1301-1302 |
| 5′05 | GTTTCTTGGAGTACTCTACGTC | 3′045 | TGTTCCAGTACTCGGCGCT | 171 | 1303-1305 |
| 5′03 | TACTTCCATAACCAGGAGGAGA | 3′17 | CCCGCCTGFACTTCCAGGAA | 200 | 1305 |
| 5′05 | GTTTCTTGGAGTACTCTACGTC | 3′11 | TCTGCAATAGGTGTCACACT | 224 | 1401, 1404, 1405 |
| 5′08 | ACTACTCTACGGGTGAGTGTT | 1407, 1408 | |||
| 5′03 | TACTTCCATAACCAGGAGGAGA | 3′12 | GTAGGTGTCCACCGCGGCCCG | 146 | 1402, 1404, 1305, |
| 1306, 1409 | |||||
| 5′04 | GTTTCTTGGAGCAGGTTAAACA | 3′19 | CTGTTCCAGTCTCCGCGA | 188 | 1410 |
| 5′52.1 | TTTATTGGAGGCTGCGTAAGTC | 3′13 | CTGTTCCAGGACTCGGCGA | 171 | DRB3 |
| 5′52.2 | GTTTCTTGGAGCTGCTTAAGTC | 3′14 | GCTGTTCCAGTAACTCGGCAT | 173 | 0101-0301 |
| 5′53 | GAGCGAGTGTGGAACCTGA | 3′048 | CTGCACTGTGAAGCTCTCCA | 213 | DRB4 |
| 0101 | |||||
| 5′51 | GTTTCTTGCAGCCAGGATAAGTA | 3′01 | CCGCGCCTGCTCCAGGAT | 198 | DRB5 |
| 3′16 | CCGCGGCGGCCTGTC | 207 | 0101-0202 | ||
| TGCCAAGTGGAGCACCCAA | GCATCTTGCTCTGTGCAGAT | 796 | Human growth hormone |
HLA-DRB1 allele-specific of PCR/SSP amplification production.
| Portion (lane) | HLA DRB1 allele | Primer |
|---|---|---|
| 1 | 0101-0102 | 5′01+3′047+3′048 |
| 2 | 0103 | 5′01+3′10 |
| 3 | 1501-1502 | 5′02+3′01 |
| 4 | 1601-1602 | 5′02+3′02 |
| 5 | 0301-0302 | 5′03+3′03 |
| 6 | 0301 | 5′06+3′048 |
| 7 | 0302-1302-1305 | 5′03+3′047 |
| 1402-1403-1409 | ||
| 8 | 0401-0414-1401 | 5′04+3′047+3′048 |
| 9 | 0701-0702 | 5′07+3′079 |
| 10 | 0809-0806 | 5′08+3′05 |
| 11 | 0901 | 5′09+3′079 |
| 12 | 1001 | 5′10+3′047 |
| 13 | 1101-1105 | 5′05+3′06 |
| 14 | 1201-1202 | 5′08+3′08 |
| 15 | 1301-1302-1306 | 5′03+3′10 |
| 16 | 1303-1304-1305 | 5′05+3′045 |
| 17 | 1305 | 5′03+3′17 |
| 18 | 1401-1404-1405 | 5′05+5′08+3′11 |
| 1407-1408 | ||
| 19 | 1402-1305-1306 | 5′03+3′12 |
| 20 | 1410 | 5′04+3′19 |
| 21 | DRB3 | 5′52.1+5′52.2+3′13+3′14 |
| 0101-0301 | ||
| 22 | DRB4 0101 | 5′53+3′048 |
| 23 | DRB5 0101-0201 | 5′51+3′01+3′16 |
Conventional proteinase K-phenol-chloroform method was applied to extract genome DNA from peripheral blood lymphocyte (PBL). The DNA concentration was determined by UV spectrophotometer and adjusted to 100ng/μL subsequently.
Every PCR reaction tube contains 60ng genomic DNA to be tested, 0.5U Taq polymerase, 200μmol of dNTP, 2pmol SSP of each HLA-DRB1 alleles. Conditions for PCR: denaturation at 94°C for 5.5min; renaturation at 55°C for 40s; elongation at 72°C for 1min, 30 cycles. In the last cycle, the elongation step was extended to 2min at 72°C. PCR product was analyzed by 2% agarose gel (ethidium bromide was contained) electrophoresis (10–15V/cm gel, 20min). Bromchlorophenol blue was used as an indicator. DNA band appeared obviously under UV. 1–20 pairs of primers were DRB1 allele-specific or group-specific, respectively. 21–23 pairs of primers were HLA-DRB3, -DRB4, DRB5 gene-specific, and they were related to different DRB1 alleles, respectively. Human growth hormone (796bp) was used as positive control of PCR amplification, and the assessment of PCR results referred to references.15
Direct counting method was used to calculate allele GF. Hardy-Weinberg equilibrium test was used to analyze samples, and relative risk (RR) was calculated compared with control group. The formula: gene frequency (GF)=detected number of people with HLA-DRB1 alleles/total number of people in each experimental group; RR=a×d/b×c, where a and b represent number of patients with positive (+) HLA-DRB1 alleles, and number of patients with negative (−) HLA-DRB1 alleles in the same group, respectively; c and d represent number of healthy people with positive (+) HLA-DRB1 alleles, and number of healthy people with negative (−) HLA-DRB1 alleles in the same group, respectively.
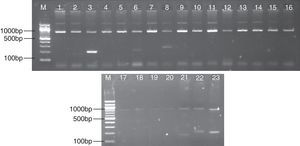
The results of PCR products of each HLA-DRB1 allele are shown in Fig. 1. The distribution of HLA-DRB1 alleles and RR was analyzed. According to Table 3, in drug use patients compared with healthy controls, the frequencies of *0401-041 (p<0.05, RR=9.22), *1001 (p<0.05, RR=19.27) were significantly higher; in HIV-infected patients compared with healthy controls, the frequencies of HLA-DRB1 *0101-0103 (Pc<0.01, RR=6.42), *0401-0411 (Pc<0.01, RR=21.93), *1001 (Pc<0.05, RR=10.43) were significantly higher, indicating the relationship with HIV susceptivity, while the frequencies of DRB1 *1501-1502 (Pc<0.01, RR=0.06), *1101-1105 (Pc<0.01, RR=0.16), *1301-1302 (Pc<0.01, RR=0.10), DRB4 (Pc<0.01, RR=0.11), DRB5 (Pc<0.01, RR=0.10) were significantly lower than that in healthy controls, indicating the relationship with HIV repellency; in HIV-infected drug users compared with healthy controls, the frequencies of HLA-DRB1 *0101-0103 (p<0.01, RR=7.26), *0401-0411 (p<0.01, RR=16.73), *0801-0806 (p<0.01, RR=5.49), *1001 (p<0.05, RR=22.91). *1401/1404/1405 (p<0.05, RR=5.41) were significantly higher, while the frequencies of DRB1 *1301/1302 (p<0.01, RR=0.600649), 1501-1502 (p<0.01, RR=0.18), DRB5 (p<0.01, RR=0.40) were significantly lower. This study showed that in HIV-infected group and HIV-infected drug use group, the number of alleles whose frequency changed significantly was significantly higher than that in drug use group, indicating that variability of drug use combined with HIV infection among different individuals is closely related with HLA-DRB1 gene polymorphism.
PCR amplification results of HLA-DRB1 Alleles. Positive bright band appeared in Lanes 3, 6, 8, representing the allelotypes of HLA-DRB1 were HLA-DRB1*1501-1502, 0301, 0401-0414-1401. And Lanes 21, 22, 23 represent DRB3, DRB4, and DRB5, respectively. Each lane includes an internal PCR control amplicon of human growth hormone gene (about 800bp concordant with theoretical values).
Distribution of HLA-DRB1 alleles in each experimental group.
| DRB alleles | Control group | Drug user group | HIV infected group | Drug user with HIV infection group | Comparison between Drug user group and Control group | Comparison between HIV infected group and Control group | Comparison between Drug user with HIV infection group and Control group | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Noa | Rate | Noa | Rate | Noa | Rate | Noa | Rate | X12 | Pc1 | RR1 | X22 | Pc2 | RR2 | X32 | Pc3 | RR3 | |
| DRB1 | |||||||||||||||||
| 0101-0103 | 4 | 0.019 | 1 | 0.042 | 7 | 0.109 | 5 | 0.122 | 0.547 | p>0.05 | 2.27 | 8.35 | p<0.01 | 6.42 | 10.708 | p<0.01 | 7.26 |
| 0401-0411 | 1 | 0.004 | 1 | 0.042 | 6 | 0.094 | 3 | 0.073 | 3.524 | p<0.05 | 9.22 | 12.44 | p<0.01 | 21.93 | 10.400 | p<0.01 | 16.73 |
| 0809-0806 | 9 | 0.042 | 2 | 0.0833 | 4 | 0.063 | 8 | 0.195 | 0.822 | p>0.05 | 2.06 | 0.451 | p>0.05 | 1.51 | 12.866 | p<0.01 | 5.49 |
| 1001 | 1 | 0.004 | 2 | 0.0833 | 3 | 0.047 | 4 | 0.098 | 10.278 | p<0.01 | 19.27 | 3.545 | p<0.05 | 10.43 | 15.365 | p<0.01 | 22.91 |
| 1401/1404/1405 | 2 | 0.009 | 1 | 0.0417 | 1 | 0.016 | 2 | 0.048 | 1.798 | p>0.05 | 4.59 | 0.179 | p>0.05 | 1.67 | 3.442 | p<0.05 | 5.41 |
| 1501-1502 | 47 | 0.221 | 3 | 0.125 | 1 | 0.016 | 2 | 0.049 | 1.186 | p>0.05 | 1.19 | 14.441 | p<0.01 | 0.06 | 6.524 | p<0.01 | 0.18 |
| DRB5 | 72 | 0.038 | 5 | 0.208 | 3 | 0.047 | 7 | 0.171 | 1.514 | p>0.05 | 0.52 | 21.129 | p<0.01 | 0.10 | 4.491 | p<0.01 | 0.40 |
| 1601-1602 | 8 | 0.038 | 2 | 0.083 | 2 | 0.031 | 2 | 0.049 | 1.12 | p>0.05 | 2.33 | 0.056 | p>0.05 | 0.83 | 0.114 | p>0.05 | 1.31 |
| 0301- | 18 | 0.085 | 2 | 0.083 | 5 | 0.078 | 4 | 0.098 | 0.00385 | p>0.05 | 0.985 | 0.026 | p>0.05 | 0.92 | 0.0741 | p>0.05 | 1.17 |
| 0302- | 6 | 0.028 | 2 | 0.083 | 2 | 0.031 | 1 | 0.024 | 2.01 | p>0.05 | 3.14 | 0.017 | p>0.05 | 1.11 | 0.0183 | p>0.05 | 0.86 |
| 0302-, 1302- | 17 | 0.080 | 4 | 0.17 | 3 | 0.047 | 3 | 0.073 | 2.01 | p>0.05 | 3.14 | 0.797 | p>0.05 | 0.57 | 0.0209 | p>0.05 | 0.91 |
| 0701-0702 | 10 | 0.047 | 3 | 0.125 | 6 | 0.094 | 3 | 0.073 | 2.53 | p>0.05 | 2.9 | 1.981 | p>0.05 | 2.10 | 0.487 | p>0.05 | 1.60 |
| 0901 | 29 | 0.14 | 2 | 0.083 | 11 | 0.721 | 6 | 0.15 | 0.529 | p>0.05 | 0.577 | 0.508 | p>0.05 | 1.32 | 0.0301 | p>0.05 | 1.09 |
| 1101-1105 | 36 | 0.17 | 2 | 0.083 | 2 | 0.031 | 5 | 0.12 | 0.233 | p>0.05 | 1.29 | 7.891 | p<0.01 | 0.16 | 0.563 | p>0.05 | 0.68 |
| 1201-1202 | 3 | 0.014 | 1 | 0.042 | 4 | 0.063 | 2 | 0.0488 | 0.989 | p>0.05 | 3.04 | 2.942 | p>0.05 | 4.67 | 2.14 | p>0.05 | 3.59 |
| 1301-1302 | 28 | 0.131 | 2 | 0.0833 | 1 | 0.016 | 1 | 0.0244 | 0.452 | p>0.05 | 0.601 | 7.047 | p<0.01 | 0.10 | 3.90 | p<0.05 | 0.17 |
| 1305- | 7 | 0.033 | 2 | 0.0833 | 2 | 0.031 | 3 | 0.0731 | 1.503 | p>0.05 | 2.68 | 0.004 | p>0.05 | 0.95 | 1.48 | p>0.05 | 2.32 |
| 1402, 1404, 1409 | 12 | 0.056 | 3 | 0.125 | 3 | 0.047 | 5 | 0.122 | 1.715 | p>0.05 | 2.39 | 0.086 | p>0.05 | 0.82 | 2.37 | p>0.05 | 2.33 |
| DRB3 | 25 | 0.117 | 5 | 0.208 | 5 | 0.078 | 4 | 0.0976 | 1.614 | p>0.05 | 1.98 | 0.785 | p>0.05 | 0.63 | 0.13 | p>0.05 | 0.81 |
| DRB4 | 81 | 0.038 | 9 | 0.375 | 4 | 0.063 | 14 | 0.341 | 0.00255 | p>0.05 | 0.98 | 0.063 | p<0.01 | 23.366 | 0.22 | p>0.05 | 0.84 |
| 1410 | 2 | 0.09 | 1 | 0.042 | 1 | 0.016 | 1 | 0.0244 | 1.798 | p>0.05 | 4.59 | 0.179 | p>0.05 | 1.67 | 0.663 | p>0.05 | 2.63 |
| 1303-1304 | 25 | 0.117 | 1 | 0.042 | 3 | 0.047 | 2 | 0.0488 | 1.266 | p>0.05 | 0.33 | 2.692 | p>0.05 | 0.37 | 1.703 | p>0.05 | 0.39 |
Drug users become one of the major sources responsible for HIV transmission in China. According to “China HIV Prevention and Protection Report (2011)”, by the end of October 2011, the estimated number of HIV patients in China is 78,000. Among these patients, 40% of them were infected from needle injection. According to sentinel surveillance data, 7% of needle injection drug users obtained a positive outcome for HIV test in recent years. Once HIV-1 intrudes into human body, virus will integrate with cells and can barely be removed in their lifetime. Since viral genes can change into various types, viral genome is far more complicated than any of a recognized virus. Therefore, HIV-1 infection has become a serious threat to human health. Two reasons lead to the wide dissemination of HIV in drug users1: high-risk drug use behaviors. Generally, sharing injectors is considered to be the main reason for HIV infection2; unsafe/commercial sexual behavior of drug users was another reason contributing to the popularity of AIDS among drug users. In addition, some studies showed that except for drug dependence and virus, different immune reactions among different individuals was important in the development of disease among drug use combined with HIV infectors. HLA has a close relationship with human immune functional status, and 39.8% genes of the 128 HLA functional genes are related to immunity.15 Especially, almost every gene in class II shows related functions to immunity. Among them, HLA-DRB1 (contains 494 alleles) polymorphism is the most complicated. This polymorphism is the main genetic factor for immune response in different individuals among the group and differences in susceptibility to disease.
To date, many studies have showed the close relationship between HLA-DRB1 gene polymorphism and infectious diseases. As a main gene cluster to regulate immune response, HLA complex is closely related with antiviral immune response. Some special HLA genotypes may have effect on the strength of the immune response after the viral infection, thus leading to the persistent virus infection. You et al. studied alleles HLA-DRB1 *1301/1302, 1501/1502, 1201/1202 in patients with hepatitis B using PCR-SSP technology, and drew a conclusion that allele GF of HLA-DRB1 *1301/1302 among patients with chronic hepatitis B was 0.56%, which was significantly lower than control group (p<0.05); the other two alleles had no significant difference (p>0.05). Cheng et al. found that allele GF of HLA-DRB1 *1201/1202 was significantly higher than control group (p<0.001, RR=4.99) among patients with liver cirrhosis in Hubei province, while GF of HLA-DRB1 *1501/1502 alleles was significantly lower (p<0.005, RR=0.30). Other researchers found that allele GF of HLA-DRB1 *1301/1302 among European chronic hepatitis B infectors was 5.7%, which was lower than healthy people (26.7%); they also found that allele GF of HLA-DRB1 *1301/1302 was significantly higher among patients with acute hepatitis B than that among patients with chronic hepatitis B.16–18 HLA-DRB1 gene polymorphism also has important research value in HIV infection. MacDonald et al.19 found that in Africa Kenya population, genotype of HLA-DRB1 *0102 was a protective gene of HIV, while HLA-A *2301 is associated with a significantly increased risk of HIV-1 infection. Among American white people, HLA-DQB1 *0602 is related to susceptivity of HIV, while among African-Americans, HLA-DQB1 *0605 is the allele which is associated with susceptivity. Moreover, American white people with HLA-DQB1 *0603 shows certain degree of repellency, but African-Americans with HLA-DRB1 *04 shows repellency to HIV infection.20
Our findings showed that allele frequency of HLA-DRB1 *0101-0103, *0401-0411, *0801-0806, *1001, *1401/1404/1405 in HIV-infected drug use group were significantly higher than that in control group (p<0.01 or p<0.05), indicating that they may be the predisposing genes of drug use combined with HIV infection, while allele frequency of HLA-DRB1 *1301/1302, *1501-1502, DRB5 were significantly lower than that in control group, indicating that they may be the protecting genes. On the contrary, HLA-DRB1 *0101-0103, * 0401-0411, *1001 may related to HIV susceptivity in HIV-infected group, while HLA-DRB1 *1501-1502, *1101-1105, *1301-1302, DRB4, DRB5 may related to HIV repellency. Compared with control group, allele frequency of only HLA-DRB1 *0401-0411, *1001 were significantly higher. Studies showed that different alleles have different frequencies, thus leading to distinguished difference of susceptivity and repellency to disease among different groups. Only two allele's frequency changed significantly in drug users group, but there were 3 susceptibility alleles and 5 resistance alleles in HIV-infected group; 5 susceptibility alleles and 3 resistance alleles in drug user with HIV-infected group. These studies indicated the close relationship between HLA-DRB1 alleles and HIV infection. Susceptivity or repellency of drug use combined with HIV is different in different races. Thus, the investigation of HLA-DRB1 alleles has certain clinical and basic effect on the susceptivity or repellency of drug use combined with HIV. The study on the relationship between HLA and drug use combined with HIV at DNA level will help for elucidating pathogenesis of diseases and mechanism of immune response, and meanwhile will help for finding new effective target in the treatment of drug use combined with HIV infection.
The preliminary study is limited for the relatively small number of patients studied. Further study will be studied including more patients, and also focused on clinicopathologic parameters in a larger cohort of patients. Besides, the association of HLA-DRB1 alleles and susceptibility of HIV infection will be studied by comparing the HIV infected individuals considering the drug use as variable in our further research.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by the National Natural Science Foundation of China (Grant No. 30972754) and the Fundamental Research Funds for the Central Universities.