Tuberculosis (TB) is one of the infectious diseases with high mortality in the world. DNA amplification techniques have been used to overcome barriers to the diagnosis of this disease. However, the success of these methodologies is highly dependent on the DNA obtained from the sample. This study was carried out to verify whether the DNA extracted by sonication (in house method) could yield suitable DNA for amplification by real-time PCR (qPCR). Sixty sputum samples were submitted to DNA extraction using sonication compared to a commercial method (Detect-TB kit, Labtest/MG-Brazil). All DNA samples were amplified by qPCR for IS6110 region (IS6110-qPCR/SYBR Green assay). Out of 60 samples, 40 were positive for TB; of these, all had positive results when extracted by sonication (100%) and 80% when extracted by the commercial method. The limit of detection (LOD) of Mycobacterium tuberculosis (H37Rv strain) by qPCR was 14 CFU/mL when the DNA was extracted by sonication, compared to countless colonies when extracted by commercial kit. In conclusion, the sonication protocol (without purification step) proved to be a simple, fast, and suitable method for obtaining DNA for use in qPCR from sputum samples.
Tuberculosis (TB) is one of the main causes of death and the leading cause from a single infectious agent worldwide.1 Millions of people continue to fall sick with TB each year,2 more than 1.7 billion people (about 25 percent of the world population) are estimated to be infected with M. tuberculosis. In 2017, 10 million individuals became ill with TB and 1.6 million died.3
Early diagnosis is one of the strategies to combat the disease.4 Methodologies based on the amplification of nucleic acids by Polymerase Chain Reaction (PCR) for the detection of M. tuberculosis DNA have improved the diagnosis and can overcome problems associated with classical methods.5 The use of molecular techniques, besides contributing to detect the increase in the number of TB cases, also allows to quickly characterize resistance and define the existing genotypes in a given community.6
However, differences in sensitivity and specificity when using PCR techniques are a reality. There are many reasons for this variability. One is the applied DNA extraction protocol.7 The DNA extraction method must provide a good yield and DNA of quality, elimination of PCR inhibitors, implementable as a laboratory technique, resulting in a genetic material of good quality for additional molecular analyses, while still being simple and fast.8 Many commercial methods are available (QIAGEN QIAmp DNA mini kit, Nucleic Acid and Protein Purification-Macherey, Nagel, DynaMagTM-2 Magnet, Detect-TB kit), whereas DNA purification with silica, magnetic beads, Chelex beads, among others, have been described to improve DNA quality.7,9,10 Also, many laboratories use in-house methods for this step, mainly to curb costs.11–13
The DNA extraction method that uses the cellular disruption by sonication has also been used in several laboratories,14–16 but there is still doubt about the efficacy of DNA extracted directly from sputum clinical samples (which have a wide variety of microorganisms, many impurities, and dependence on good collection) for use in PCR, as the protocol does not include a purification step.17,18 Therefore, this study aimed to compare a sonication technique as the method for obtaining DNA from sputum samples with the commercial method (silica resin - Detect-TB kit, marketed by Labtest / MG-Brazil). IS6110 – qPCR/SYBRGreen was used as a confirmatory method for detecting bacterial DNA.19
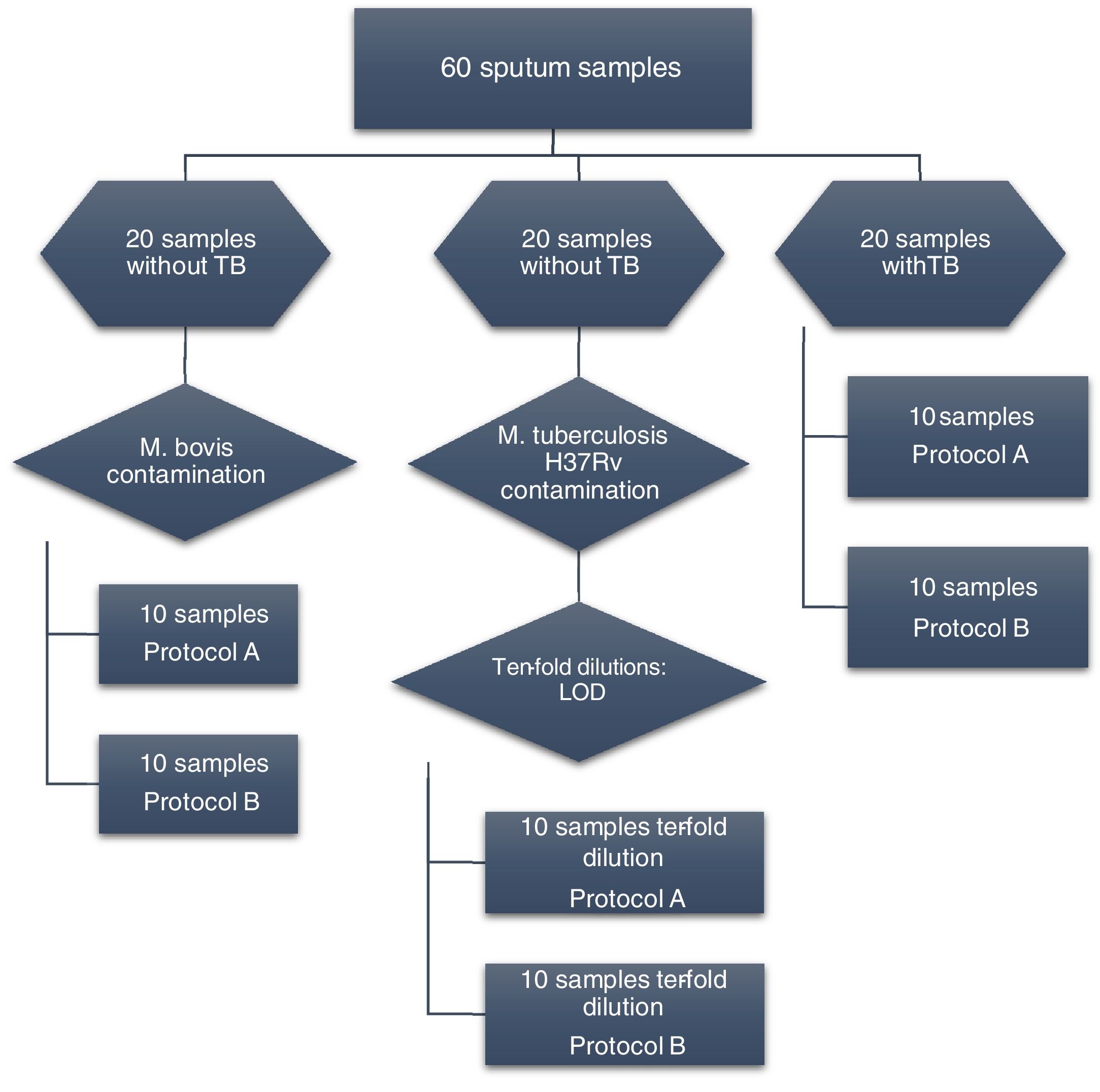
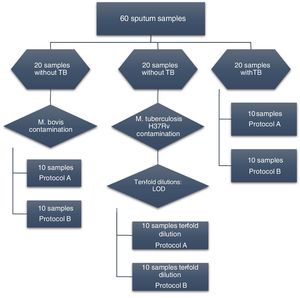
Materials and methodsSamplesSixty sputum samples were collected from patients who attended the Department of Tisiology and Leprosy in Canoas, Southern Brazil. Initially, to evaluate DNA extraction with the two protocols from the same bacterial concentration, 20 sputum samples known to be from TB-negative patients were contaminated with 25 μL (100 CFU/mL) of an aqueous suspension of Mycobacterium bovis strain.
qPCR amplification was verified in 20 sputum samples from patients with tuberculosis. To determine the limit of detection, 20 negative samples were contaminated with 25 μL of 10-fold dilutions of M. tuberculosis reference strain H37Rv.
The sputum samples used in this study were also assessed on smear slides, culture, and GeneXpert MTB/RIF (Cepheid, EUA). The flowchart of the analysis is presented in Fig. 1.
Sample preparationThe sputum samples were decontaminated and treated with 4% sodium hydroxide (NaOH) and PBS (Phosphate-buffered saline) solution containing sodium chloride, potassium chloride, sodium phosphate, monobasic potassium phosphate, and ultrapure water, at an adjusted pH of 7.4. Briefly, a total of 500 μL of sputum was transferred to a 1.5 mL microtube and added the corresponding volume of NaOH 4% solution, followed by vortex homogenization, and incubation at 37 °C for 15 min. Subsequently, it was centrifuged 3000 g for 15 min and the supernatant was discarded. The pellet was resuspended in 500 µL of PBS buffer and homogenized on a vortex.
DNA extractionProtocol A - silica resin (Detect-TB kit, Labtest / MG-Brazil)The silica resin method was described by Michelon et al. (2011)20 as an adaptation of the Boom et al. protocol (1990).21 Briefly, the protocol consisted of a cell lysis step with a solution containing 8 M guanidine hydrochloride, pH 6.4, 40 mM EDTA and 2 g/dL polyoxyethylene octyl phenyl ether, followed by incubation in a thermoblock at 100 °C for 10 min, aiming at the release and solubilization of the genomic DNA. Purification of the released DNA was carried out through specific uptake by a silica resin in the presence of chaotropic agents. Subsequently, a wash sequence was performed with distinct solutions: the first containing guanidine hydrochloride, the second containing buffered sodium chloride concentrate at pH 7.0 and polyoxyethylene sorbitan monolaurate 0.1%, and the third containing concentrated sodium chloride and buffered at pH 7.5 with polyoxyethylene 0.1% monolaurate. After washing, the purified DNA was eluted in 25 μL of ultrapure water and frozen at −20 °C until use.
Protocol B - sonication (in-house method)A microtube containing 500 µL of sputum sample pretreated as mentioned above, was placed at 95 °C for 20 min. After this step, it was placed in the sonication bath for 15 min, and then it was centrifuged for 5 min at 3000 g. Subsequently, 100 μL of the supernatant (containing the DNA) was transferred to a new microtube and frozen at −20 °C until use.
Quantitative assessment of extracted DNAThe DNA quantification was performed by spectrophotometry. The DNA absorbance and concentration were evaluated for nucleic acid purity (ideal ratio: A260/A280 ≥ 1.8 and A260/A230 = 2). Samples were quantified in the SpectraMaxPlus® (Eppendorf) spectrophotometer using 2 μL of DNA according to the manufacturer's instructions.
Limit of detection (LOD)Colonies of M. tuberculosis H37Rv strain grown in Ogawa Kudoh culture medium were scraped and placed in a glass tube containing beads to solubilize and homogenize bacterial colonies, and added ultrapure water (the volume can be stipulated by the operator). The turbidity of the bacterial solution was compared to the turbidity of the Macfarland number 1 scale (widely used in microbiology, especially for conventional antibiograms). Ten-fold dilutions (turbidity scale 1 solution) was performed. From these dilutions, 25 μL were mixed to 20 TB-negative sputum samples. The same volume of 25 μL of the dilutions was inoculated in 7H10 culture medium, and the CFUs was assessed for each dilution.
Real-time PCR (IS6110 – qPCR/SYBR Green)For evaluation of the viability of the extracted DNA, IS6110 real-time PCR was performed with the Step One Real-Time PCR Systems (AB Applied Biosystems) equipment and the amplified products were detected using the SYBR® Green (fluorophore). The reaction was standardized in a final volume of 20 μL containing 10 μL of Fast SYBR® Green Master Mix, 4 pmol of each primer (INS1 5 'CGTGAGGGCATGGAGGTGGC 3′ and INS2 5 'GCGTAGGCGTCGGTGACAAA 3′) and 5 μL of extracted DNA and quantified. The conditions of the amplification were as follows: activation of the enzyme at 95 °C for 20 s, denaturation at 95 °C for 1 s, and finally annealing and extension at 62 °C for 20 s. For each run, a negative reaction control (PCR mix containing ultrapure water) and a positive control reference (M. tuberculosis H37Rv - 80 ng/μL) were used. Amplification and dissociation curves were analyzed with StepOne software version 2.3. For the analysis of the amplification curve, the parameters "threshold" (detectable fluorescence level) and "baseline" (defined limit of PCR cycles) were used to determine the Ct (cycle threshold) of each sample. The dissociation curve was used to observe the possible amplification of non-specific products or primer dimers. For standardization of the ideal concentration of primers at 4 pmol, it was used the analysis of the dissociation curve of known samples and the analysis of amplicons in 1.5% agarose gel, stained with ethidium bromide.
Standard curve and efficiencyThe setup of the standard curve was performed according to the manufacturer's recommendations (AB Applied Biosystems-Thermofisher Scientific)—five points and in duplicate—for assessing DNA amplification protocol. Ten-fold dilutions were performed from the quantified DNA of M. tuberculosis strain H37Rv (using as a genomic marker the element of insertion IS6110 of the bacterium, by qPCR). The PCR mixture contained 5 μL of DNA from each dilution for each reaction, and 15 μL of the mix, totaling the final volume of 20 μL per reaction. Efficiency, calculated using StepOne software version 2.3, was of 96.15% (reference value: 90–100%), R² of 0.97 (indicates the closeness of fit between the linear regression line of the standard curve and the data of individual Ct points), −3.418 of slope (indicates the efficiency of amplification by PCR assay) and Y-inter of 16.87 (indicates the expected Ct for the sample).
Ethical aspectsThe study was approved by the Lutheran University of Brazil (ULBRA) research ethics committee (approval number 2011-340H).
Statistical analysisAll data were analyzed using IBM® SPSS® statistics (version 21.0). The Shapiro-Wilk test was used for the analysis of type of data distribution. Student's T-test for independent samples and Mann-Whitney test were used to identify possible differences of DNA concentrations and Cts obtained between the two protocols evaluated. All tests performed were interpreted using a two-sided significance level of 5% (p < 0.05).
ResultsThe qPCR amplification of the DNA extracted from the 20 contaminated samples with M. bovis (10 DNAs extracted per protocol) showed that DNA extracted by Protocol A (silica resin) was detected in 8 of the 10 samples (80.0%) and all 10 DNAs extracted by Protocol B (sonication) were amplified (100%).
The spectrophotometric quantification of the DNAs amplified and purity evaluation by the different protocols are shown in (Table 1). Regarding DNA purity at wavelength 260/280 nm, significant differences were found between the results of two protocols (1.67 ± 0.35 vs. 0.97 ± 0.07; p < 0.01). At the wavelength of 260/230 nm, a statistical difference was detected between the data evaluated, Protocol A differed from Protocol B (0.14 ± 0.12 vs. 0.74 ± 0.33; p < 0.01). There was a significant difference in DNA concentration between the two protocols used in the analysis (58.83 ng/µL ± 76.65 ng/µL vs. 340.43 ng/µL ±154.34 ng/µL; p < 0.01).
Comparison between the averages of the degrees of purity and DNA concentrations obtained by the two protocols applied to clinical samples contaminated with M. bovis strain.
| Protocols | A260/280 (≥1.8) | A260/230 (<2.0) | DNA ng/µL | Intensity amplicons electrophoresis gel |
|---|---|---|---|---|
| A | 1.67 ± 0.35 | 0.14 ± 0.12 | 58.83 ± 76.65 | ++ |
| B | 0.97 ± 0.07 | 0.74 ± 0.33 | 340.43 ± 154.34 | +++ |
| p-values | <0.01* | <0.01* | <0.01* |
Protocols: A) Detect-TB Kit; B) Sonication.
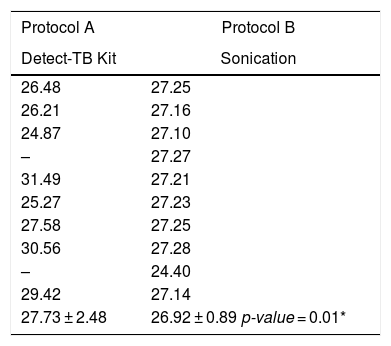
The amplifications Cts by real-time PCR and the means of each DNA extraction protocol are shown in (Table 2). Comparing Cts of protocol 1 vs. 2, there was statistical difference (27.73 ± 2.48 vs. 26.92 ± 0.89; p < 0.01).
CTs corresponding to the extraction protocols applied from DNA extracted from clinical samples contaminated with M. bovis strain.
| Protocol A | Protocol B |
|---|---|
| Detect-TB Kit | Sonication |
| 26.48 | 27.25 |
| 26.21 | 27.16 |
| 24.87 | 27.10 |
| – | 27.27 |
| 31.49 | 27.21 |
| 25.27 | 27.23 |
| 27.58 | 27.25 |
| 30.56 | 27.28 |
| – | 24.40 |
| 29.42 | 27.14 |
| 27.73 ± 2.48 | 26.92 ± 0.89 p-value = 0.01* |
Regarding the LOD (using ten-fold dilutions of the M. tuberculosis strain H37Rv), with protocol B the qPCR assay detected up to 14 CFUs (1: 1000 dilution), whereas with Protocol A the LOD was countless colonies.
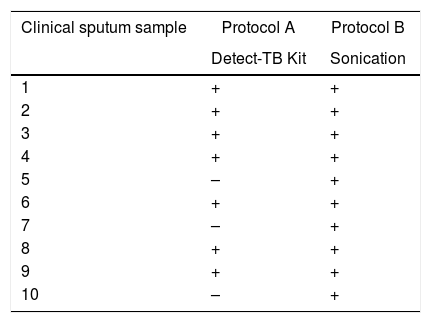
Among the 20 samples evaluated from patients with TB, DNA extracted with Protocol A was detected TB in seven of the 10 (70.0%) samples and in all 10 (100%) DNA extracted with Protocol B (Table 3).
DNA purity and quantification extracted from M. tuberculosis sputum samples followed the same standards as those of M. bovis contaminated samples. Table 4 shows the mean purity and quantification of the nucleic acid extracted with the two protocols. Regarding DNA purity at the wavelength of 260/280 nm, there was significant difference between the two protocols (2.19 ± 1.62 vs. 1.09 ± 0.21; p < 0.01). When analyzing DNA purity at wavelength 260/230 nm, there was no significant difference between the two protocols (0.07 ± 0.04 vs. 0.68 ± 0.50; p < 0.01). Concerning DNA quantification, there was a significant difference (20.75 ng/µL ± 11.74 ng/µL vs. 37.01 ng/µL ± 36.91 ng/µL; p < 0.01) between the protocols used in the analysis.
Comparison between the averages of the degrees of purity and the DNA concentrations obtained by protocols applied to clinical samples of patients with TB.
| Protocols | A260/280 (≥1.8) | A260/230 (<2.0) | DNA ng/µL | Intensity amplicons electrophoresis gel |
|---|---|---|---|---|
| A | 2.19 ± 1.62 | 0.07 ± 0.04 | 20.75 ± 11.74 | ++ |
| B | 1.09 ± 0.21 | 0.68 ± 0.50 | 37.01 ± 36.91 | +++ |
| p-values | <0.01* | <0.01* | <0.01* |
Protocols: A) Detect-TB Kit; B) sonication.
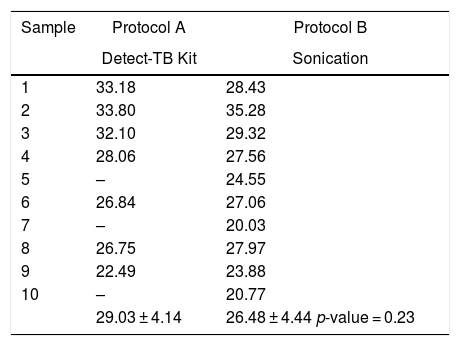
The results of the Cts for the DNA extracted samples from patients with TB using the two protocols are shown in Table 5. The difference observed between Protocol A vs. B was not significant (29.03 ± 4.14 vs. 26.48 ± 4.44; p = 0.23) when considering the samples with Mycobacterium DNA detected.
CTs corresponding to the extraction protocols applied from DNA extracted from clinical samples from patients with TB.
| Sample | Protocol A | Protocol B |
|---|---|---|
| Detect-TB Kit | Sonication | |
| 1 | 33.18 | 28.43 |
| 2 | 33.80 | 35.28 |
| 3 | 32.10 | 29.32 |
| 4 | 28.06 | 27.56 |
| 5 | – | 24.55 |
| 6 | 26.84 | 27.06 |
| 7 | – | 20.03 |
| 8 | 26.75 | 27.97 |
| 9 | 22.49 | 23.88 |
| 10 | – | 20.77 |
| 29.03 ± 4.14 | 26.48 ± 4.44 p-value = 0.23 |
We assessed in this study if extraction DNA from sputum samples (simple and low-cost technique) with sonication would provide DNA suitable for the detection of the M. tuberculosis by qPCR, through the identification of the insertion sequence IS6110, being reported as the most specific and widely explored genetic marker (number of IS6110 copies per genome might influence the sensitivity of the test).22,23 Sputum is the most common biological sample used for the diagnosis of pulmonary TB disease.24 As compared to urine and blood, sputum is thicker and more viscous but provides better insight into TB and other respiratory-related diseases.25 When used directly for DNA extraction and subsequent detection by PCR, it is known that sputum samples, present barriers related to the presence of internal Taq inhibitors, as well as presence of several microorganisms that are part of the microbiota of the respiratory and oral tracts.26–28 Thus, sample preparation and choice of an efficient method for DNA extraction is one of the main challenges for the molecular diagnosis of TB.29
Regarding the use of the available commercial kits, it is known that they are generally preferred because they provide greater reproducibility, quality control, and automation potential.30 Silica matrices (Detect-TB kit, marketed by Labtest/MG-Brazil - Protocol A) provide efficient and selective binding of nucleic acids (NA), having as characteristic the procedure of purification of DNA.31 The mechanism involved in this technique is the affinity between negatively charged NA and positively charged silica material, resulting in selective binding of NA.32 In clinical applications, even using kits, to obtain high yields of NA is still challenging, since sputum samples often exhibit a low final concentration of DNA, which is masked against a large background of contaminants.
In our study, there was variability in some standards regarding the quantitative evaluation of the extracted DNA. Nonetheless, Protocol B provided a higher amount of extracted DNA, lower Cts in the plot amplification (by qPCR) and profiles of fragments with a more uniform aspect, when visualized in electrophoresis. In line with our findings, the study by Krinitsina et al. (2015)33 showed that the purity values and some DNA concentrations measured by absorbance, clearly differ from the visible data of real-time PCR and electrophoresis detection (fluorescence intensity of amplicons). Depending on the extraction method applied, it is important to point out that purification of DNA molecules is known to be influenced by their specificities of size, nucleotide composition, topology, and association with proteins.30 We believe that reagent residues with different chemicals, or the need for further washes during the application of the protocols, may have influenced the absorbance reading. Therefore, a quantitative evaluation of the extracted DNA cannot be evaluated as an absolute parameter.
According to the study by Aldous et al. (2005),34 it was possible to conclude that the purification step using silica resin was not necessary for the extraction of DNA from sputum samples before the PCR technique for identification of M. tuberculosis since it makes the method more extensive and impairs laboratory routine. Comparing the use of Kit (Detect-TB – Protocol A) with in-house method (Sonication – Protocol B), it was concluded that the DNA extracted by Kit presented higher purity, but the amount of extracted DNA was, in many cases, inconsistent for detection.32 In a study by Psifidi et al. (2015),35 11 different protocols were tested for DNA extraction and further used in genomic and microarray sequencing. Among them, the one that obtained the shortest execution time, lowest cost, and the best results were the in-house protocol, as our results also demonstrated.
The analyses by Liu et al. (2013),36 Greenly, and Tester (2015)37 showed that the use of ultrasound allows cell walls to be cleaved by shock waves, created by rapid changes in pressure caused by sonication. The study by Granger et al. (2004)15 reported that the use of sonication improved mycobacterial DNA by removing background bacteria and other cells. In addition to these experiments, studies by Pchelintsev, Adams, and Nelson (2016)17 showed that the use of sonication minimizes the risk of cross-contamination between samples. Dai et al. (2016)16 concluded that the sonication-based technique is considered more promising, efficient, and economical for homogeneous cell decomposition, in a short period.
Regarding the limit of detection (LOD) of qPCR/SYBR Green assay, Protocol B provided a good detection capacity (up to 14 CFU/mL), even from smaller amounts of DNA (1:1000 dilution), consumed fewer reagents, with fewer execution steps, proving to be effective, less laborious, and fast method that presented greater potential for application in sputum samples.8 The LOD obtained in our study was similar to that found in the study by Chakravorty et al. (2017),38 which analyzed the performance of Xpert MTB/RIF Ultra as a suitable point-of-care (POC) assay for TB diagnosis, and obtained a LOD of 15.6 CFU/mL.
We recognize potential study limitations. However, our findings are important for guiding the choice of DNA extraction method applicable to clinical sputum samples. Future studies may consolidate our findings, thus allowing greater understanding of the need for NA purification for molecular detection of potential microorganisms that cause infectious diseases.
Conclusions and perspectivesIdentification techniques based on molecular biology have been established as potential tools for diagnosis. In light of this, this study showed that the extraction of DNA by sonication, even without presenting purification steps during the execution of the technique, provided suitable genetic material for identification by real-time PCR, presenting higher DNA concentrations, lower costs and complexity of execution. Thus, it is one of the most promising DNA extraction techniques for use in clinical sputum samples from patients, presenting high applicability to the diagnosis of TB, as well as the possible potential for molecular epidemiology analyses, such as Whole Genome Sequencing (WGS).
FundingGraziele Bello and Jonas Michel Wolf were supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior)/MEC/Brazil-Fellowship. Financing code 001.
Conflicts of interestThe authors declare no conflicts of interest.
The authors wish to thank the Postgraduate Program in Cellular and Molecular Biology Applied to Health - Lutheran University of Brazil (ULBRA), Center for Scientific and Technological Development (CDCT-RS), and the Department of Tisiology and Leprosy of Canoas, Brazil.